Engineering
- Why Do We Need BackendAI FastTrack 2
- Reality ML Code Is Just the Tip of the Iceberg
- Limitations of Existing MLOps Tools
- The Core Problem Fragmented ML Workflows
- The Solution BackendAI FastTrack 2 Model Serving
- A True Ally for Researchers
- RealWorld Example Migrating from Airflow to BackendAI FastTrack 2
- Airflow
- Running and Initializing the Container
- Creating a DAG
- BackendAI FastTrack 2
- Creating a new pipeline
- Uploading a file
- Adding a task
- Running a Pipeline
- Serving a Model
- Closing
- Appendix
- MNIST Dataset
Jun 29, 2025
Engineering
Backend.AI FastTrack 2 Guide for Airflow Users: Seamless Integration from Training to Serving

Jeongseok Kang
Researcher
Jun 29, 2025
Engineering
Backend.AI FastTrack 2 Guide for Airflow Users: Seamless Integration from Training to Serving

Jeongseok Kang
Researcher
Why Do We Need Backend.AI FastTrack 2?
According to a 2014 Google paper, machine learning systems inherently contain significant technical debt, and the code responsible for training and inference represents only a small fraction of the overall system. The majority of the code is dedicated to integrating various components, not just the core ML logic12.
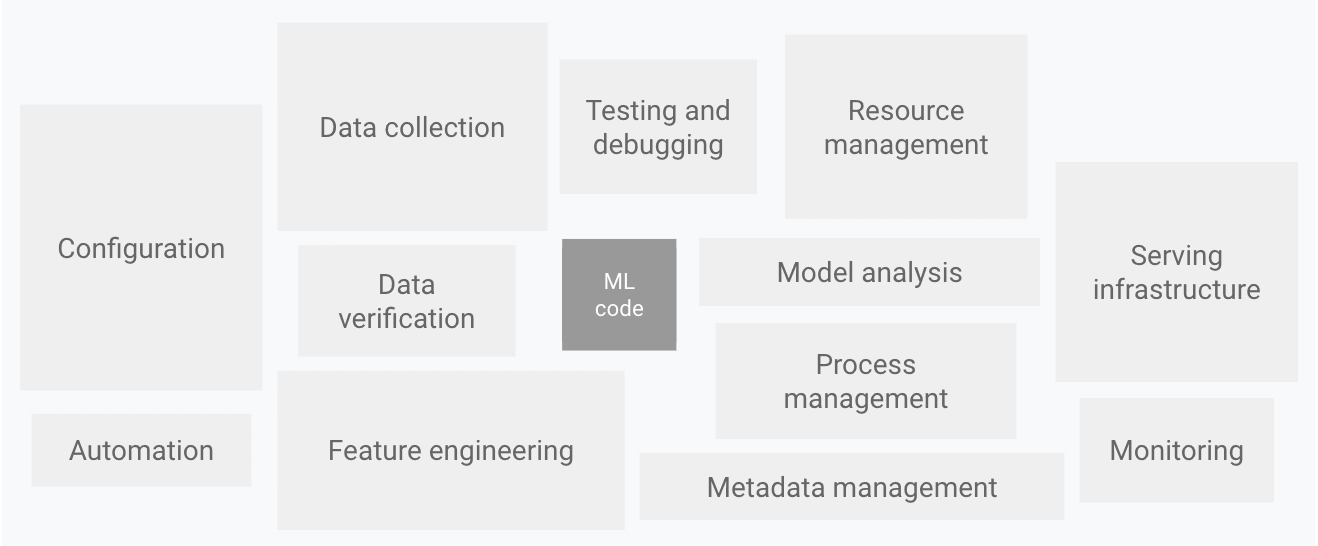
Reality: ML Code Is Just the Tip of the Iceberg
 Source: Google Cloud - MLOps: Continuous delivery and automation pipelines in machine learning
Source: Google Cloud - MLOps: Continuous delivery and automation pipelines in machine learning
The actual machine learning code is merely the visible part of a much larger iceberg, while the rest consists of infrastructure, monitoring, and data pipeline engineering. As a result, managing these dependencies and enabling systematic experimentation, monitoring, and alerting through robust MLOps pipelines is becoming increasingly important.
 Source: Machine Learning Operations (MLOps): Overview, Definition, and Architecture
Source: Machine Learning Operations (MLOps): Overview, Definition, and Architecture
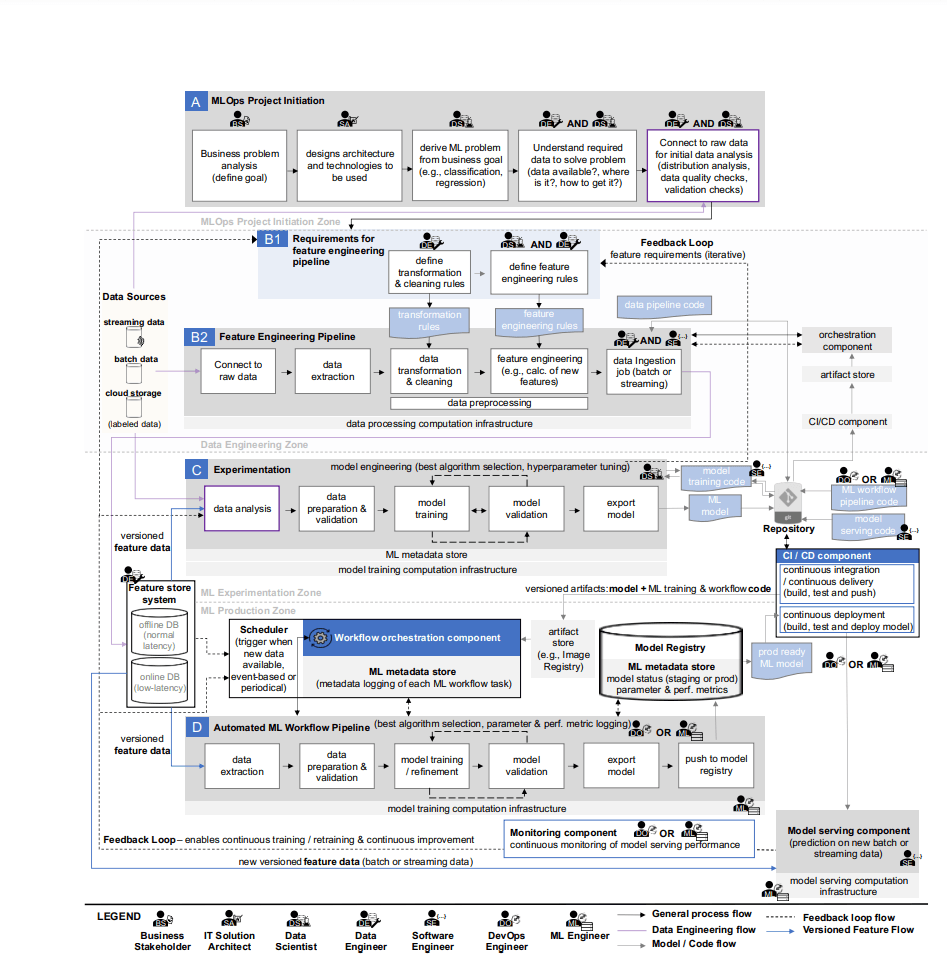
Limitations of Existing MLOps Tools
A 2023 review paper3 highlights that while open-source projects like Airflow4 and Kubeflow5 are widely used, each has its own limitations:
- Airflow: Excellent for workflow orchestration, but requires additional setup for GPU resource management and model serving.
- Kubeflow: Powerful, but demands deep Kubernetes expertise and complex configuration.
- Other tools: Often specialized for certain tasks, lacking an integrated user experience.
The Core Problem: Fragmented ML Workflows
Researchers and engineers face several challenges:
- Complex infrastructure setup: Even simple model training often requires configuring Docker, Kubernetes, environment variables, and more.
- Difficulty managing GPU resources: Efficient allocation and management of expensive GPU resources is challenging.
- Complexity of model serving: Deploying trained models as production services requires separate tools and configurations.
- Fragmented workflows: Different tools and platforms are used for each stage—training, validation, and serving—leading to inefficiencies.
The Solution: Backend.AI FastTrack 2 Model Serving
To address these challenges, Backend.AI has introduced an integrated model serving feature, as previously announced on our blog. With Backend.AI FastTrack 2, users can now leverage built-in model serving capabilities, enabling more efficient AI model deployment and management.
What is Model Serving? Model serving is the process of deploying a trained ML model as an API endpoint in a production environment, enabling real-time inference requests. This is the crucial step that transforms a model into real business value.
By integrating this feature, Backend.AI FastTrack 2 offers the following competitive advantages:
- ✅ End-to-end pipeline: Seamless workflow from model training and validation to serving.
- ✅ GPU resource optimization: Automatic scaling and efficient GPU allocation.
- ✅ Ready-to-use APIs: REST API endpoints are available immediately after training.
- ✅ Intuitive UI: Manage all tasks through a web interface.
 Source: Backend.AI Architecture
Source: Backend.AI Architecture
A True Ally for Researchers
Backend.AI FastTrack 2 allows researchers to focus on core research without getting bogged down by infrastructure complexity. There’s no longer any need to waste time on Docker configuration, Kubernetes management, or writing complex deployment scripts.

Real-World Example: Migrating from Airflow to Backend.AI FastTrack 2
In this blog, we’ll use the MNIST (Modified National Institute of Standards and Technology) classification problem as an example to demonstrate how a pipeline previously managed with Airflow can be set up more simply and efficiently with Backend.AI FastTrack 2.
Pre- and Post-Migration Comparison
| Category | Airflow | Backend.AI FastTrack 2 |
|---|---|---|
| Setup Complexity | Docker Compose, environment variables, dependency management | A few clicks in the WebUI |
| GPU Management | Manual configuration required | Automatic allocation and optimization |
| Model Serving | Requires separate tools (FastAPI, Gradio, etc.) | Integrated serving functionality |
| Operational Costs | High (complex infrastructure management) | Low (optimized resource usage) |
Airflow
Apache Airflow (hereafter Airflow) is an open-source workflow orchestration platform developed by Airbnb in 2014. It allows users to define and schedule complex data pipelines using Python code, which has led to its widespread adoption across many organizations.
The examples in this post use Airflow 3.0.2
In this blog, we’ll run Airflow using the official Docker image. If you’re not familiar with Docker environments, refer to the official Airflow documentation for detailed instructions.
Initial Airflow Setup
To use Airflow, you need several configuration files. Let’s walk through the setup process using a practical example.
[core] # https://airflow.apache.org/docs/apache-airflow/stable/configurations-ref.html#core
auth_manager=airflow.api_fastapi.auth.managers.simple.simple_auth_manager.SimpleAuthManager
simple_auth_manager_users = admin:admin,user:user
simple_auth_manager_passwords_file = ./passwords.json
dags_folder = /opt/airflow/dags
[api] # https://airflow.apache.org/docs/apache-airflow/stable/configurations-ref.html#api
host=0.0.0.0
port=8080
[api_auth] # https://airflow.apache.org/docs/apache-airflow/stable/configurations-ref.html#api-auth
jwt_secret={JWT_SECRET_KEY}
[secrets]
backend = airflow.secrets.local_filesystem.LocalFilesystemBackendPYTHONPATH=/opt/airflow/dags:$PYTHONPATH airflow standaloneservices:
airflow:
image: apache/airflow:3.0.2-python3.12
ports:
- 8080:8080
environment:
AIRFLOW_HOME: /opt/airflow
volumes:
- .:/opt/airflow
- ./dags/mnist/v1/.env:/opt/airflow/dags/mnist/v1/.env:ro
entrypoint: /opt/airflow/compose/entrypoint.shRunning and Initializing the Container
# Start Docker containers
$ docker compose -f docker-compose.yaml up -d
# Initialize (or reset) the database
$ docker exec -it <container_id> airflow db reset -yNow, you can access Airflow at the default address(http://127.0.0.1:8080)Log in using the credentials generated in passwords.json:
{"admin": "7PdAaKvPb3E3fasz", "user": "USmGxuNAvvCRuyYs"} Airflow - Login Screen
Airflow - Login Screen
After logging in, click the Dags menu on the left to see that the DAG list is currently empty.
 Airflow - DAG List Screen
Airflow - DAG List Screen
Creating a DAG
In Airflow, a DAG (Directed Acyclic Graph) is defined using Python code. Let’s create a DAG that downloads the MNIST dataset and performs model training.
import os
from datetime import datetime, timedelta, timezone
from airflow.providers.standard.operators.python import PythonVirtualenvOperator
from airflow.sdk import DAG
HF_TOKEN = os.getenv("HF_TOKEN")
with DAG(
dag_id="mnist",
default_args={
"depends_on_past": False,
"retries": 1,
"retry_delay": timedelta(minutes=5),
},
description="PyTorch MNIST classifcation example DAG",
params={
"hf_token": HF_TOKEN,
},
start_date=datetime.now(timezone.utc) + timedelta(seconds=10),
catchup=False,
tags=["example"],
) as dag:
def download_mnist_dataset(**kwargs) -> None:
import os
from pathlib import Path
import datasets
AIRFLOW_HOME = os.getenv("AIRFLOW_HOME", "/opt/airflow")
dataset_path = Path(AIRFLOW_HOME) / "datasets" / "mnist"
if not dataset_path.exists():
dataset_path.mkdir(parents=True, exist_ok=True)
dataset = datasets.load_dataset(
"ylecun/mnist",
token=kwargs.get("hf_token"),
)
dataset.save_to_disk(dataset_path)
download_mnist_dataset_task = PythonVirtualenvOperator(
task_id="download_mnist_dataset",
python_callable=download_mnist_dataset,
requirements=["datasets"],
system_site_packages=False,
op_kwargs={
"params": {
"hf_token": dag.params.get("hf_token"),
},
},
)
def train_mnist_model(**kwargs) -> None:
import os
from pathlib import Path
import torch
import torch.optim as optim
from torchvision import datasets, transforms
from torch.optim.lr_scheduler import StepLR
from mnist.v1.models import Net
from mnist.v1.trainer import Trainer
AIRFLOW_HOME = os.getenv("AIRFLOW_HOME", "/opt/airflow")
dataset_path = Path(AIRFLOW_HOME) / "datasets" / "mnist"
use_accel = not kwargs["params"]["no_accel"] and torch.accelerator.is_available()
torch.manual_seed(kwargs["params"]["seed"])
if use_accel:
device = torch.accelerator.current_accelerator()
else:
device = torch.device("cpu")
train_kwargs = {"batch_size": kwargs["params"]["batch_size"]}
test_kwargs = {"batch_size": kwargs["params"]["test_batch_size"]}
if use_accel:
accel_kwargs = {
"num_workers": 1,
"pin_memory": True,
"shuffle": True,
}
train_kwargs.update(accel_kwargs)
test_kwargs.update(accel_kwargs)
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
dataset1 = datasets.MNIST(dataset_path, train=True, download=True, transform=transform)
dataset2 = datasets.MNIST(dataset_path, train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1, **train_kwargs)
test_loader = torch.utils.data.DataLoader(dataset2, **test_kwargs)
model = Net().to(device)
optimizer = optim.Adadelta(model.parameters(), lr=kwargs["params"]["lr"])
scheduler = StepLR(optimizer, step_size=1, gamma=kwargs["params"]["gamma"])
trainer = Trainer(model)
class AttributeDict(dict):
def __getattr__(self, item):
return self[item]
for epoch in range(1, kwargs["params"]["epochs"] + 1):
trainer.train(AttributeDict(kwargs["params"]), device, train_loader, optimizer, epoch)
trainer.test(device, test_loader)
scheduler.step()
if kwargs["params"]["save_model"]:
torch.save(model.state_dict(), "mnist_cnn.ckpt")
train_mnist_model_task = PythonVirtualenvOperator(
task_id="train_mnist_model",
python_callable=train_mnist_model,
requirements=["torch", "torchvision"],
system_site_packages=False,
op_kwargs={
"params": {
"lr": 1.0,
"epochs": 14,
"batch_size": 64,
"test_batch_size": 1000,
"gamma": 0.7,
"no_accel": False,
"dry_run": False,
"seed": 1,
"log_interval": 10,
"save_model": True,
},
},
)
download_mnist_dataset_task >> train_mnist_model_taskimport torch
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
self.conv2 = nn.Conv2d(32, 64, 3, 1)
self.dropout1 = nn.Dropout(0.25)
self.dropout2 = nn.Dropout(0.5)
self.fc1 = nn.Linear(9216, 128)
self.fc2 = nn.Linear(128, 10)
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.dropout1(x)
x = torch.flatten(x, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=1)
return outputimport torch
import torch.nn.functional as F
import torch.optim as optim
class Trainer:
def __init__(self, model: torch.nn.Module) -> None:
self._model = model
def train(
self,
args,
device: torch.device,
train_loader: torch.utils.data.DataLoader,
optimizer: optim.Optimizer,
epoch: int,
) -> None:
self._model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = self._model(data)
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
if batch_idx % args.log_interval == 0:
print("Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}".format(
epoch,
batch_idx * len(data),
len(train_loader.dataset),
100. * batch_idx / len(train_loader),
loss.item(),
))
if args.dry_run:
break
def test(
self,
device: torch.device,
test_loader: torch.utils.data.DataLoader,
) -> None:
self._model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
output = self._model(data)
test_loss += F.nll_loss(output, target, reduction="sum").item()
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).sum().item()
test_loss /= len(test_loader.dataset)
print("\nTest set: Average loss: {:.4f}, Accuracy: {}/{} ({:.0f}%)\n".format(
test_loss,
correct,
len(test_loader.dataset),
100. * correct / len(test_loader.dataset),
))Then run the DAG script you created to register with Airflow.
$ docker exec -it <container_id> python dags/mnist/v1/dag.py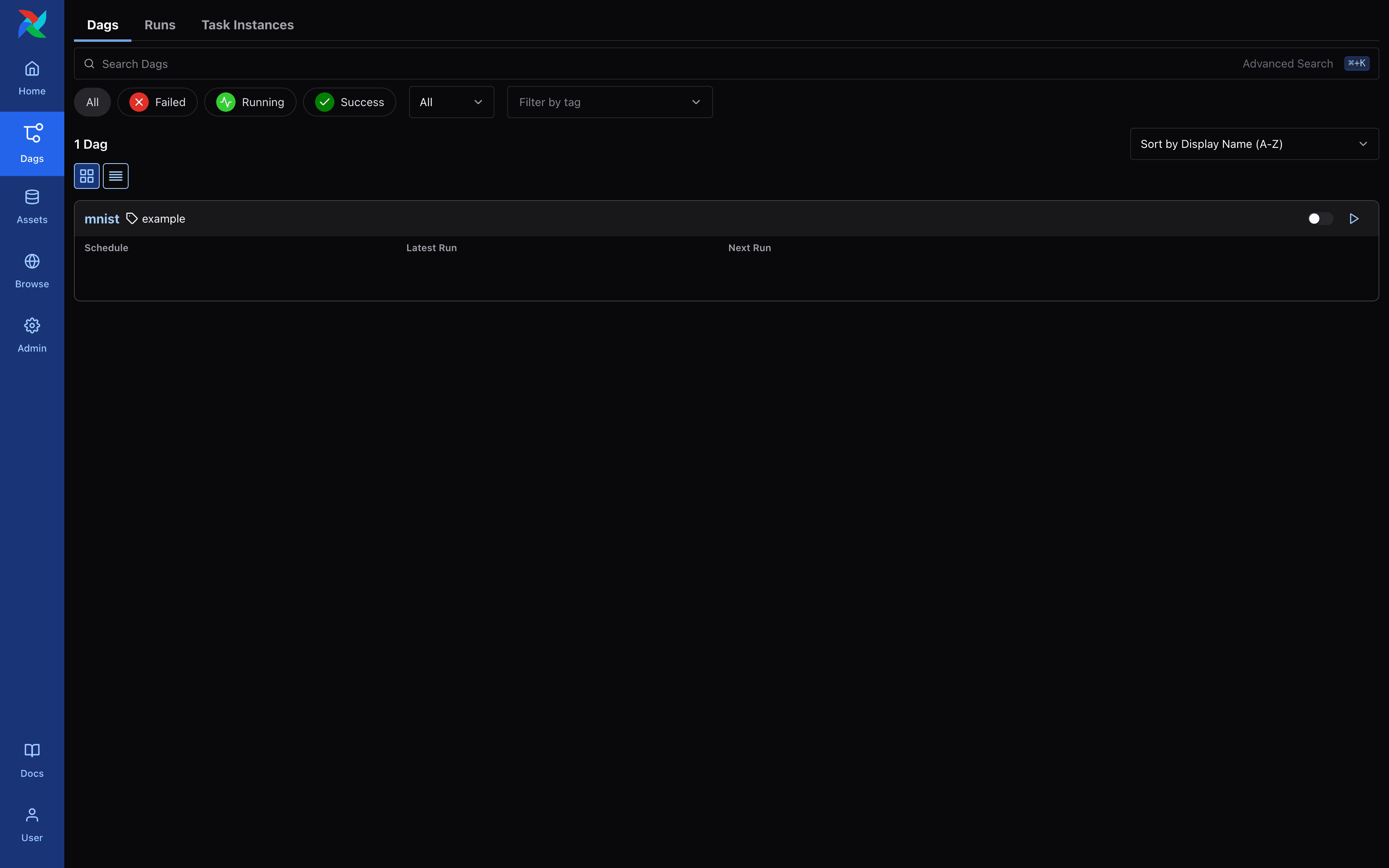 Airflow - DAG List Screen
Airflow - DAG List Screen
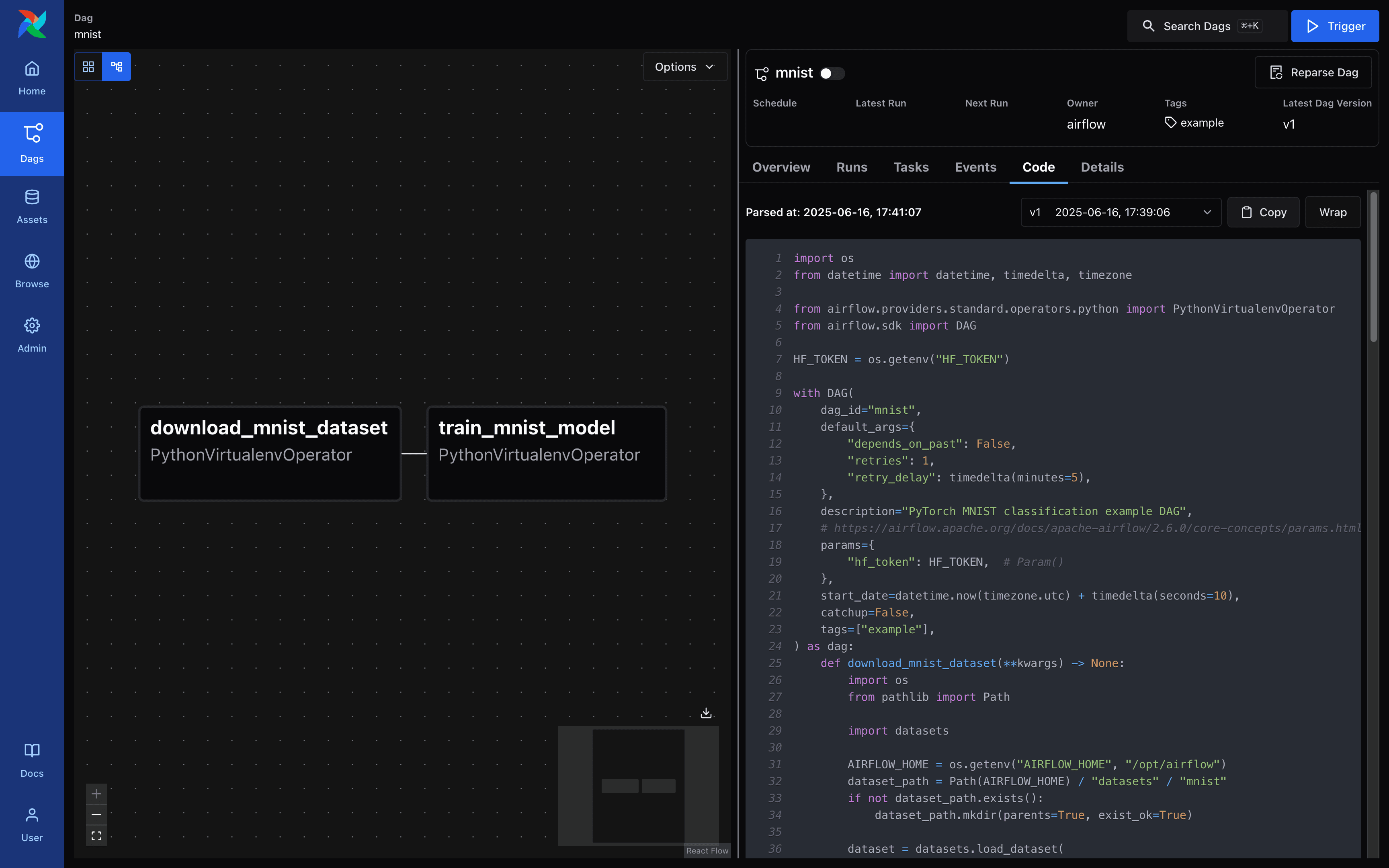 Airflow - DAG Details Screen
Airflow - DAG Details Screen
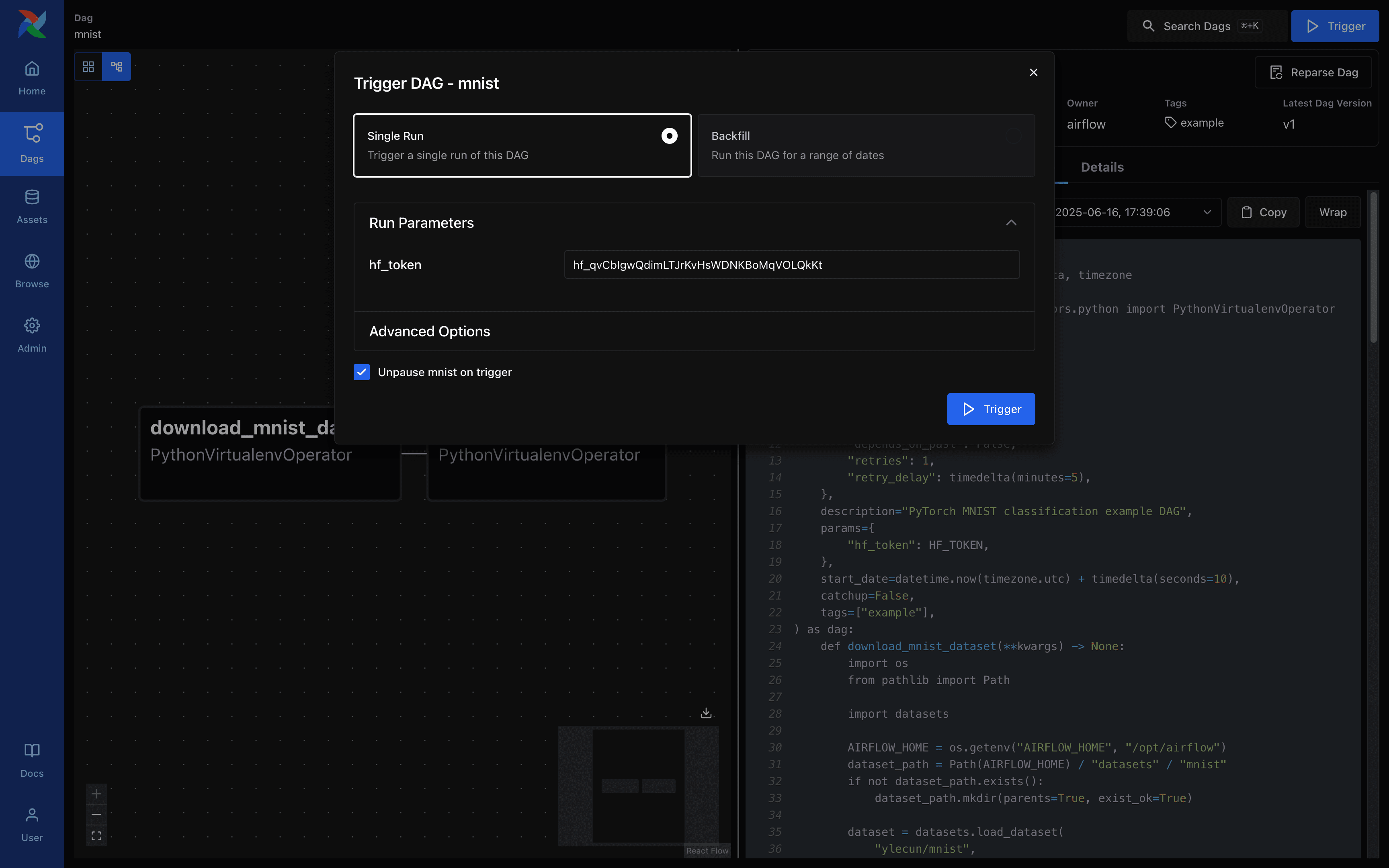 Airflow - DAG Run Request Screen
Airflow - DAG Run Request Screen
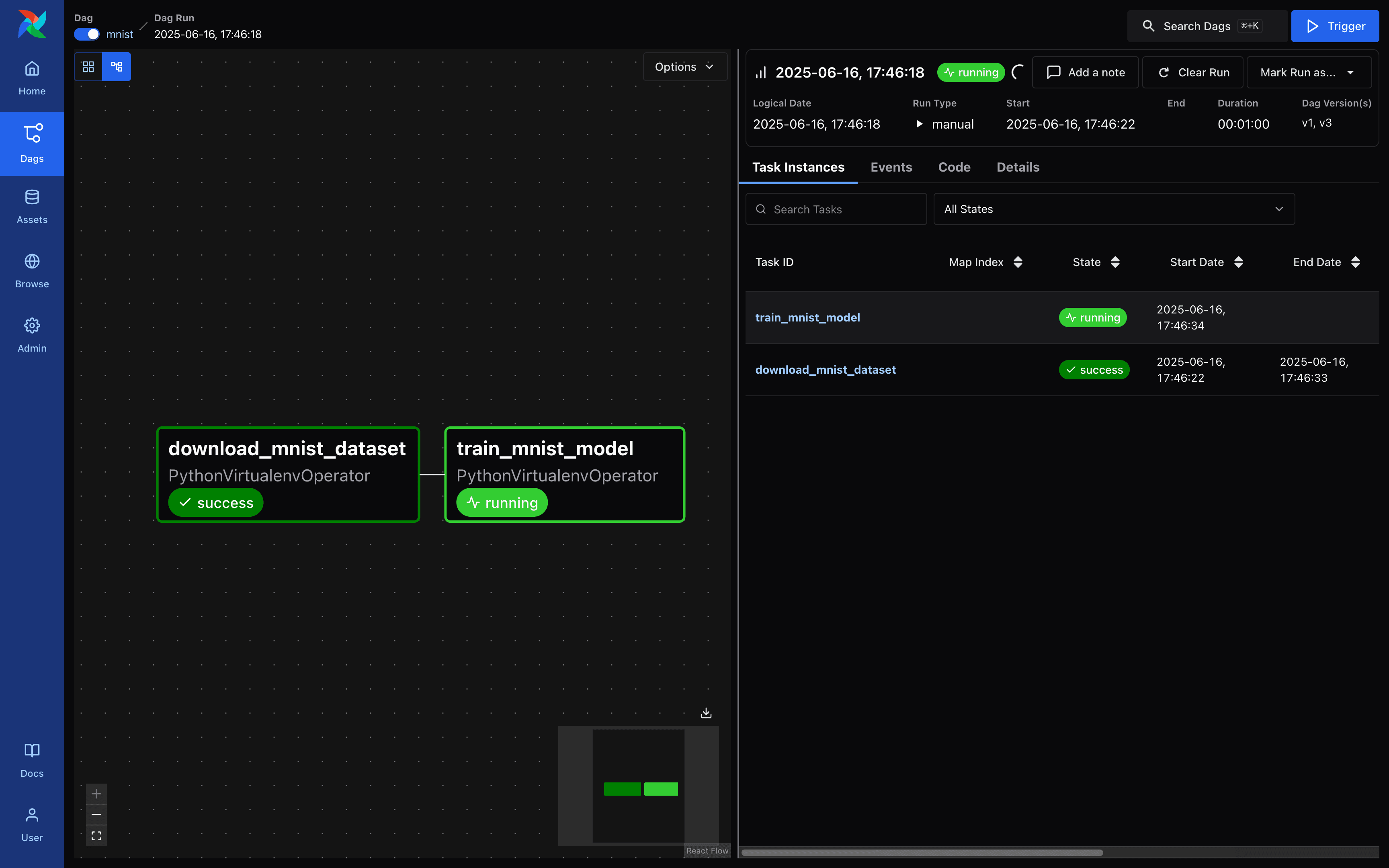 Airflow - DAG Run Request Screen
Airflow - DAG Run Request Screen
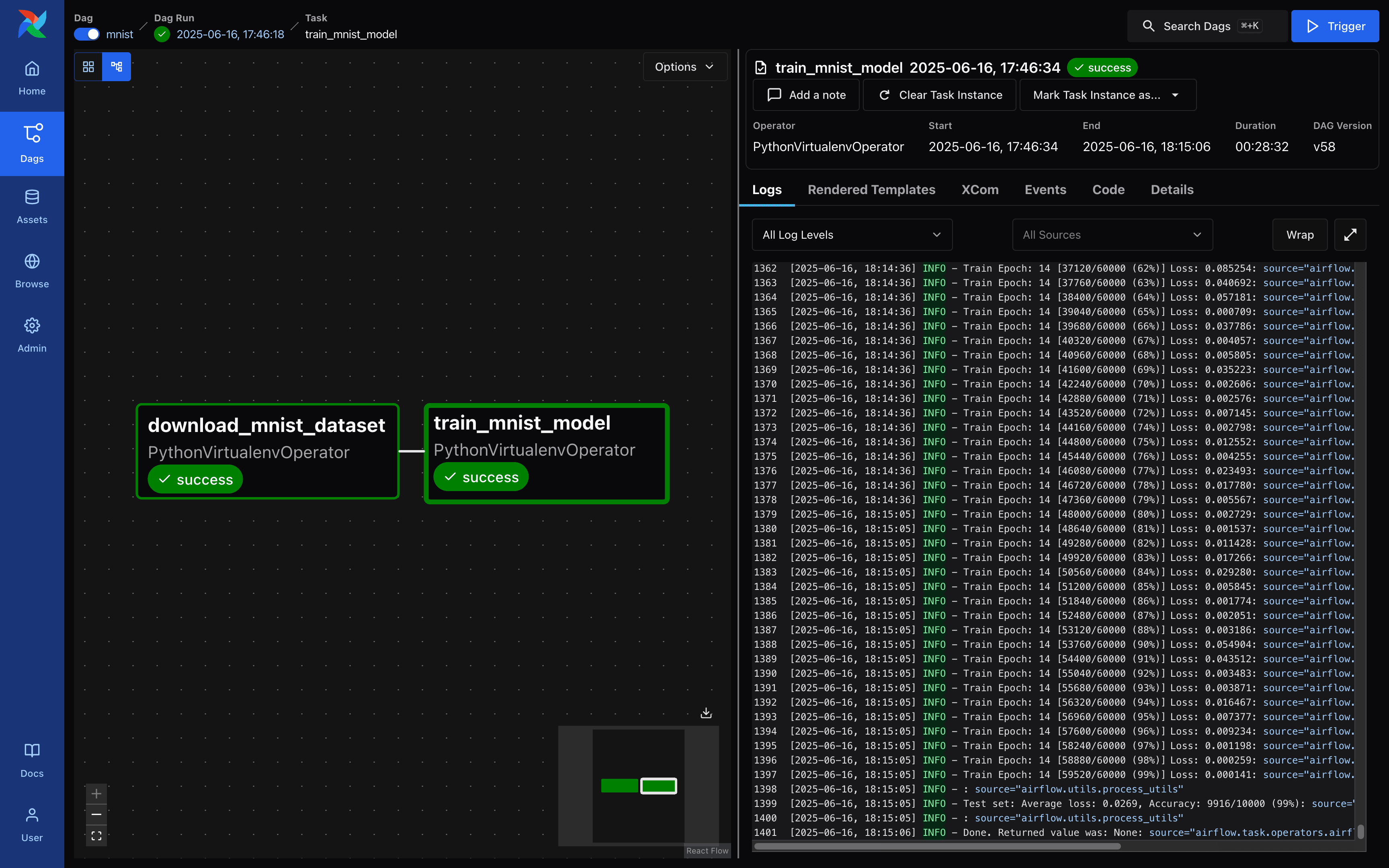 Airflow - DAG Run Request Screen
Airflow - DAG Run Request Screen
When the model is finished training, you can see that the file mnist_cnn.ckpt has been created. Next, create and run a simple Gradio demo to experiment with the MNIST model, as shown below.
$ pip install fastapi==0.115.12 gradio==5.33.0 torch==2.7.1 torchvision==0.22.1from http import HTTPStatus
from pathlib import Path
import gradio
import numpy as np
import torch
from fastapi import FastAPI, Response
from fastapi.responses import JSONResponse
from torchvision import transforms
from models import Net
model = Net()
model.load_state_dict(torch.load(Path(__file__).parent.parent.parent / "mnist_cnn.ckpt"))
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,)),
transforms.Grayscale(),
])
app = FastAPI()
@app.get("/health", status_code=HTTPStatus.OK)
async def health_check() -> Response:
return JSONResponse({"healthy": True})
def fn(img: np.ndarray) -> int:
x = transform(img).unsqueeze(0)
x = model(x)
x = x.argmax(dim=1, keepdim=True)
return int(x.item())
gradio_app = gradio.Interface(
fn=fn,
inputs=["image"],
outputs=["number"],
)
app = gradio.mount_gradio_app(app, gradio_app, path="/")$ fastapi run model-defs/gradio/main.py
FastAPI Starting production server 🚀
Searching for package file structure from directories with __init__.py files
Importing from /Users/rapsealk/Desktop/git/airflow-demo/model-defs/gradio
module 🐍 main.py
code Importing the FastAPI app object from the module with the following code:
from main import app
app Using import string: main:app
server Server started at http://0.0.0.0:8000
server Documentation at http://0.0.0.0:8000/docs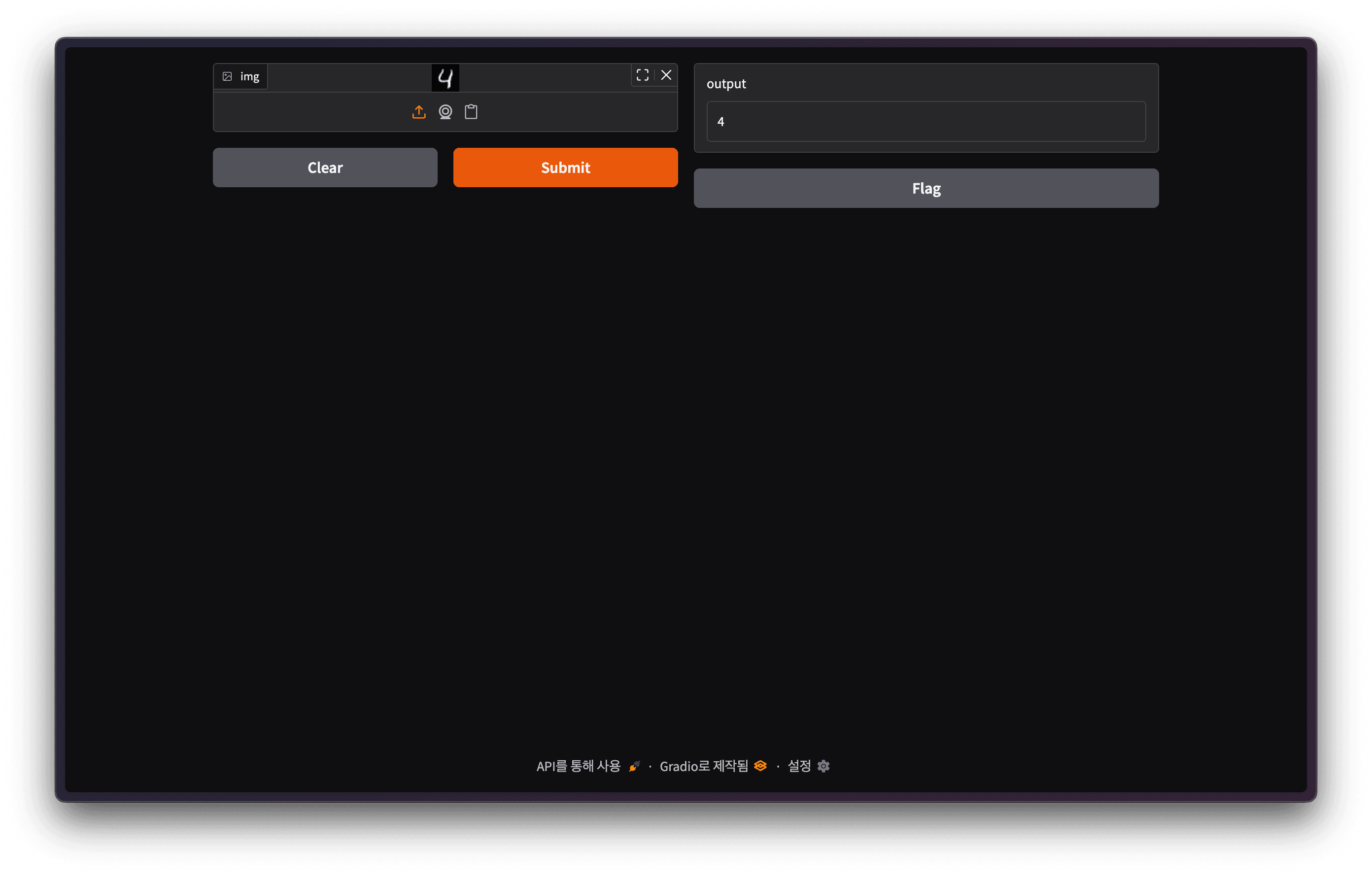 Gradio Demo Screen
Gradio Demo Screen
Backend.AI FastTrack 2
This time, we will use Backend.AI FastTrack 2 to build the MNIST training and deployment pipeline. If you are not familiar with the Backend.AI FastTrack 2 pipeline, please refer to our previous blog.
The examples contained herein were created using Backend.AI FastTrack 2 v25.9.0.
Connect to Backend.AI FastTrack 2 and click the Sign in button. You can access Backend.AI FastTrack 2 through your Backend.AI account.
 Backend.AI FastTrack 2 Login Screen
Backend.AI FastTrack 2 Login Screen
 Backend.AI Login Screen
Backend.AI Login Screen
 Backend.AI FastTrack 2 Main Screen
Backend.AI FastTrack 2 Main Screen
Creating a new pipeline
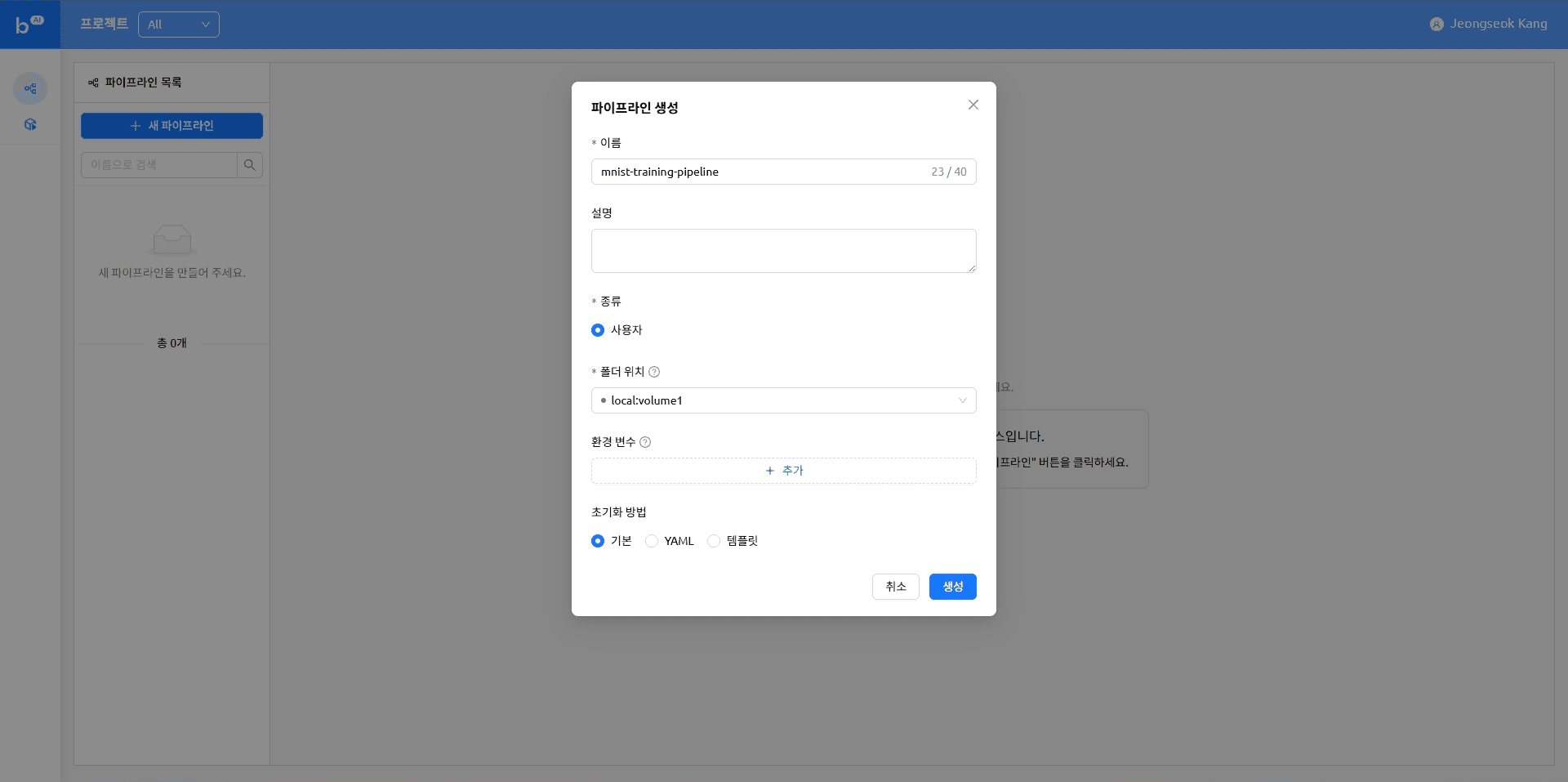
Uploading a file
The pipeline we are creating is designed to run a Python script stored in a VFolder. This table of contents covers how to upload the files needed to run the pipeline.
On the Backend.AI main screen, click the ‘Data’ item in the left menu.
 Backend.AI Main
Backend.AI Main
As you create the pipeline, you can see the pipeline folder that was created. Click a pipeline folder and upload a file.
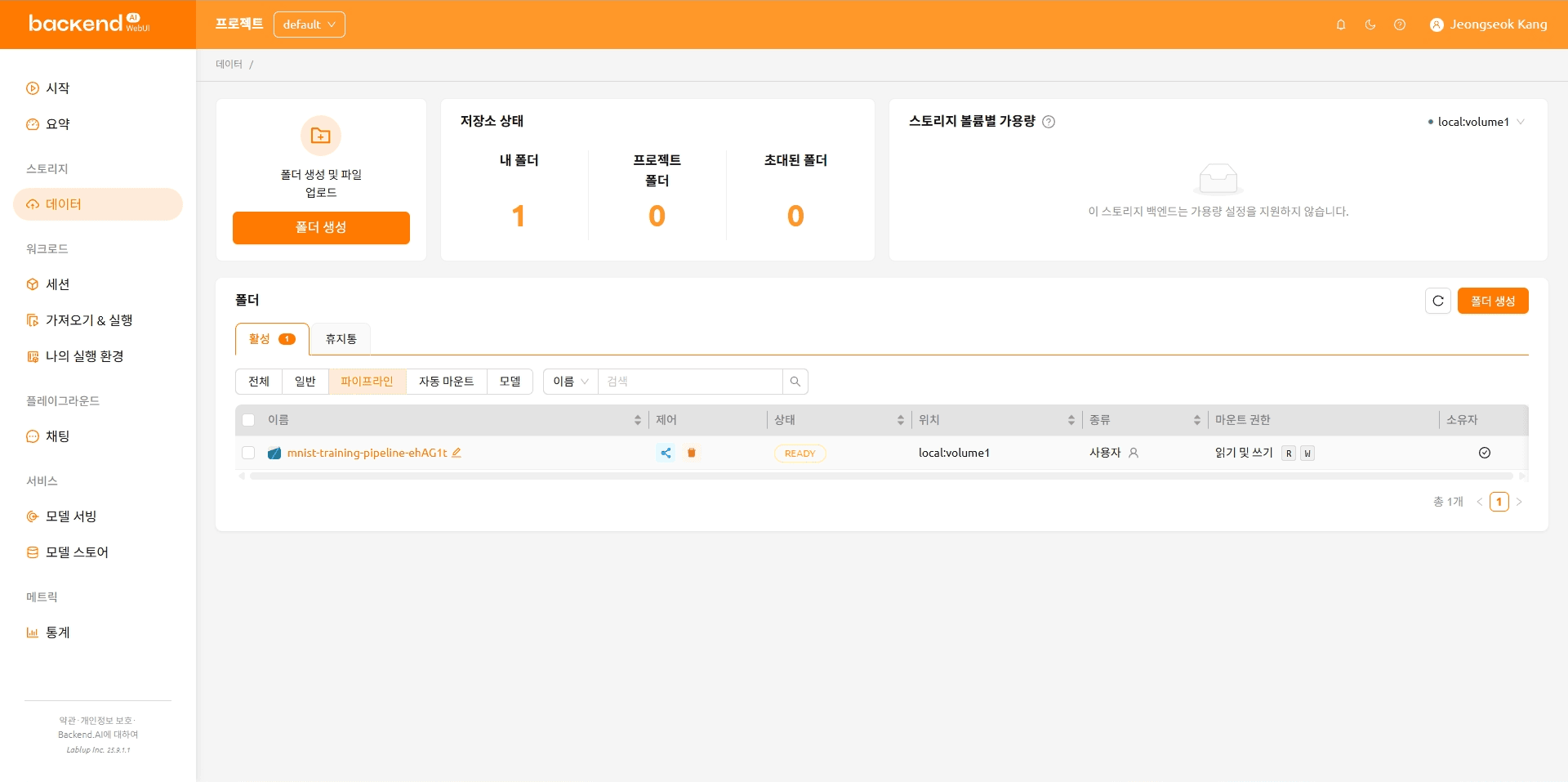 VFolder List
VFolder List
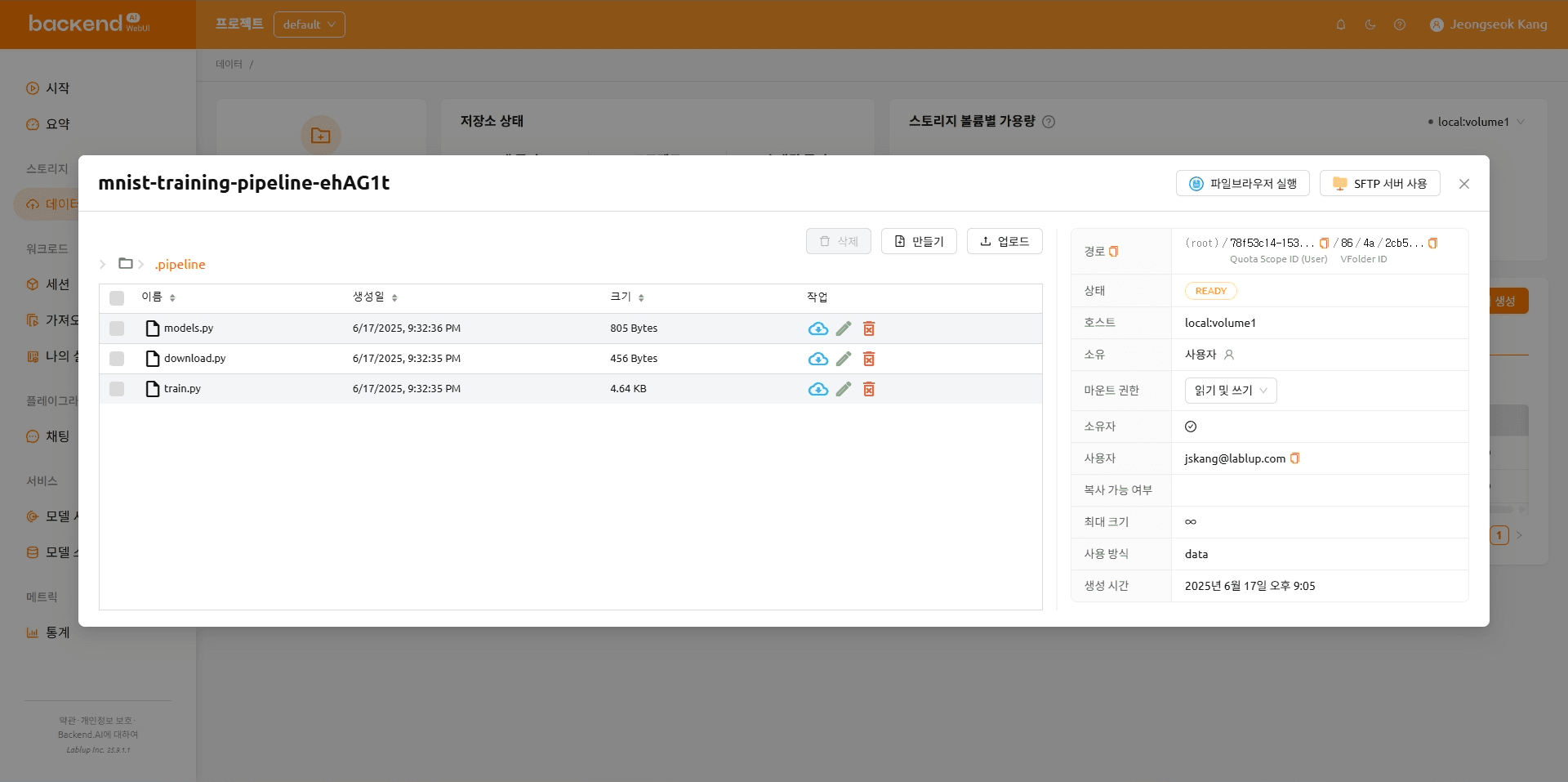 The folder after the file upload is complete
The folder after the file upload is complete
The folder organization after the files are uploaded is shown below.
mnist-training-pipeline-ehAG1t
|---- .pipeline
|---- model.py
|---- download.py
\---- train.py
import torch
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
self.conv2 = nn.Conv2d(32, 64, 3, 1)
self.dropout1 = nn.Dropout(0.25)
self.dropout2 = nn.Dropout(0.5)
self.fc1 = nn.Linear(9216, 128)
self.fc2 = nn.Linear(128, 10)
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.dropout1(x)
x = torch.flatten(x, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=1)
return outputimport os
from pathlib import Path
from torchvision import datasets
MNIST_DATASET_PATH = Path("/pipeline/vfroot") / os.getenv("MNIST_DATASET_PATH", "")
def main() -> None:
train_dataset = datasets.MNIST(MNIST_DATASET_PATH, train=True, download=True)
print(f"Train dataset is saved at: {train_dataset}")
test_dataset = datasets.MNIST(MNIST_DATASET_PATH, train=False, download=True)
print(f"Test dataset is saved at: {test_dataset}")
if __name__ == "__main__":
main()import argparse
import os
from pathlib import Path
import torch
import torch.nn.functional as F
import torch.optim as optim
from torchvision import datasets, transforms
from torch.optim.lr_scheduler import StepLR
from models import Net
MNIST_DATASET_PATH = Path("/pipeline/vfroot") / os.getenv("MNIST_DATASET_PATH", "")
CHECKPOINT_PATH = Path(os.getenv("CHECKPOINT_PATH")).joinpath(os.getenv("BACKENDAI_PIPELINE_JOB_ID", "mnist_cnn")).with_suffix(".ckpt")
def parse_args():
parser = argparse.ArgumentParser(description="Train MNIST model")
parser.add_argument("--lr", type=float, default=1.0, help="Learning rate")
parser.add_argument("--epochs", type=int, default=14, help="Number of epochs to train")
parser.add_argument("--batch-size", type=int, default=64, help="Batch size for training")
parser.add_argument("--test-batch-size", type=int, default=1000, help="Batch size for testing")
parser.add_argument("--gamma", type=float, default=0.7, help="Learning rate step gamma")
parser.add_argument("--no-accel", action="store_true", help="Disable GPU acceleration")
parser.add_argument("--dry-run", action="store_true", help="Run without saving the model")
parser.add_argument("--seed", type=int, default=1, help="Random seed for reproducibility")
parser.add_argument("--log-interval", type=int, default=10, help="Interval for logging training progress")
parser.add_argument("--save-model", action="store_true", help="Save the trained model")
return parser.parse_args()
def train(args, model, device, train_loader, optimizer, epoch):
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = model(data)
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
if batch_idx % args.log_interval == 0:
print("Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}".format(
epoch,
batch_idx * len(data),
len(train_loader.dataset),
100. * batch_idx / len(train_loader),
loss.item(),
))
if args.dry_run:
break
def test(model, device, test_loader):
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
output = model(data)
test_loss += F.nll_loss(output, target, reduction='sum').item() # sum up batch loss
pred = output.argmax(dim=1, keepdim=True) # get the index of the max log-probability
correct += pred.eq(target.view_as(pred)).sum().item()
test_loss /= len(test_loader.dataset)
print("\nTest set: Average loss: {:.4f}, Accuracy: {}/{} ({:.0f}%)\n".format(
test_loss,
correct,
len(test_loader.dataset),
100. * correct / len(test_loader.dataset),
))
def main(args) -> None:
use_accel = not args.no_accel and torch.accelerator.is_available()
torch.manual_seed(args.seed)
if use_accel:
device = torch.accelerator.current_accelerator()
else:
device = torch.device("cpu")
train_kwargs = {"batch_size": args.batch_size}
test_kwargs = {"batch_size": args.test_batch_size}
if use_accel:
accel_kwargs = {
"num_workers": 1,
"pin_memory": True,
"shuffle": True,
}
train_kwargs.update(accel_kwargs)
test_kwargs.update(accel_kwargs)
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
dataset1 = datasets.MNIST(MNIST_DATASET_PATH, train=True, download=True, transform=transform)
dataset2 = datasets.MNIST(MNIST_DATASET_PATH, train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1,**train_kwargs)
test_loader = torch.utils.data.DataLoader(dataset2, **test_kwargs)
model = Net().to(device)
optimizer = optim.Adadelta(model.parameters(), lr=args.lr)
scheduler = StepLR(optimizer, step_size=1, gamma=args.gamma)
for epoch in range(1, args.epochs + 1):
train(args, model, device, train_loader, optimizer, epoch)
test(model, device, test_loader)
scheduler.step()
if args.save_model:
CHECKPOINT_PATH.parent.mkdir(parents=True, exist_ok=True)
torch.save(model.state_dict(), CHECKPOINT_PATH)
if __name__ == "__main__":
args = parse_args()
main(args)After creating the model folder, upload the files below.
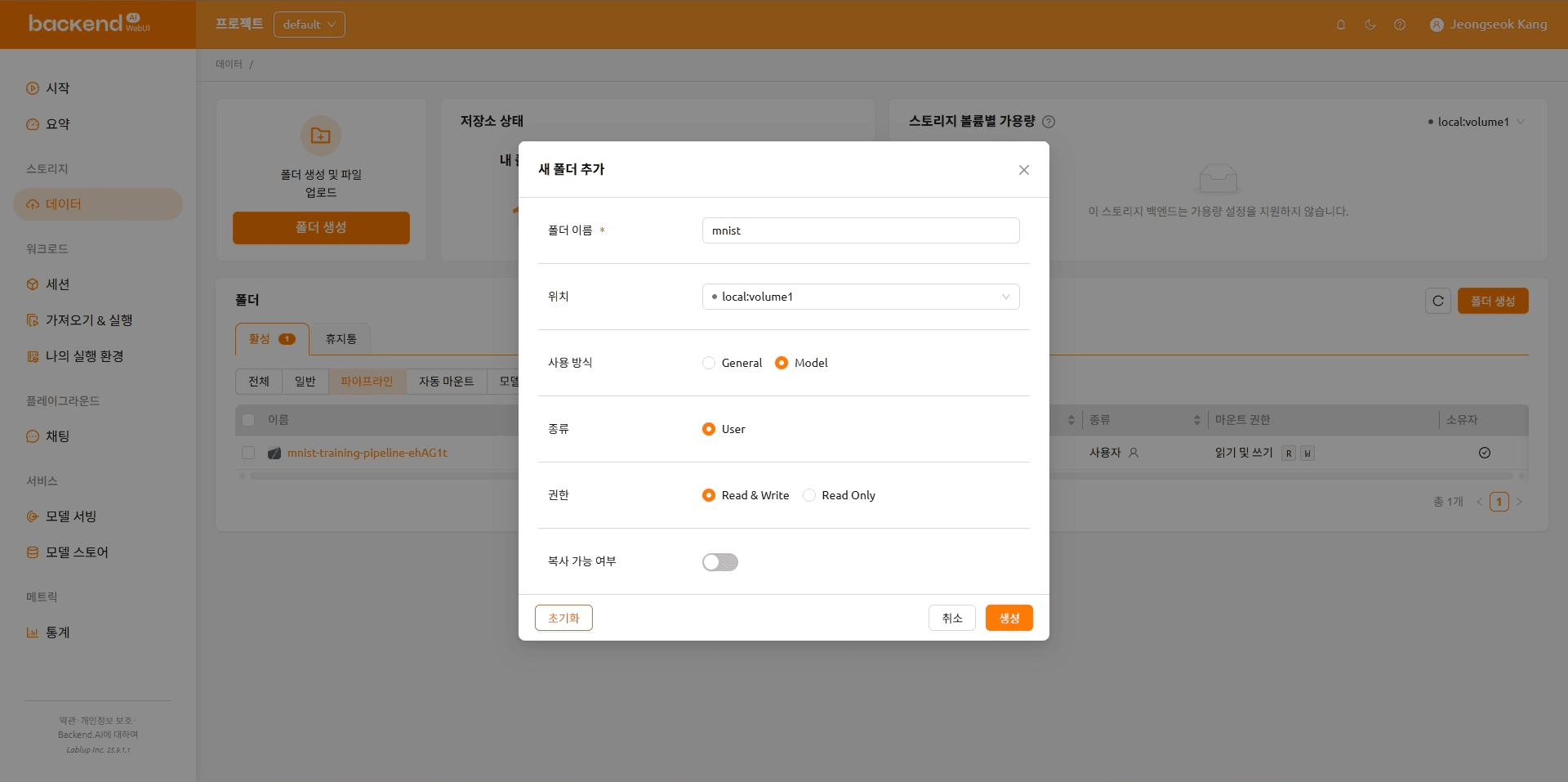 Create a Model Folder
Create a Model Folder
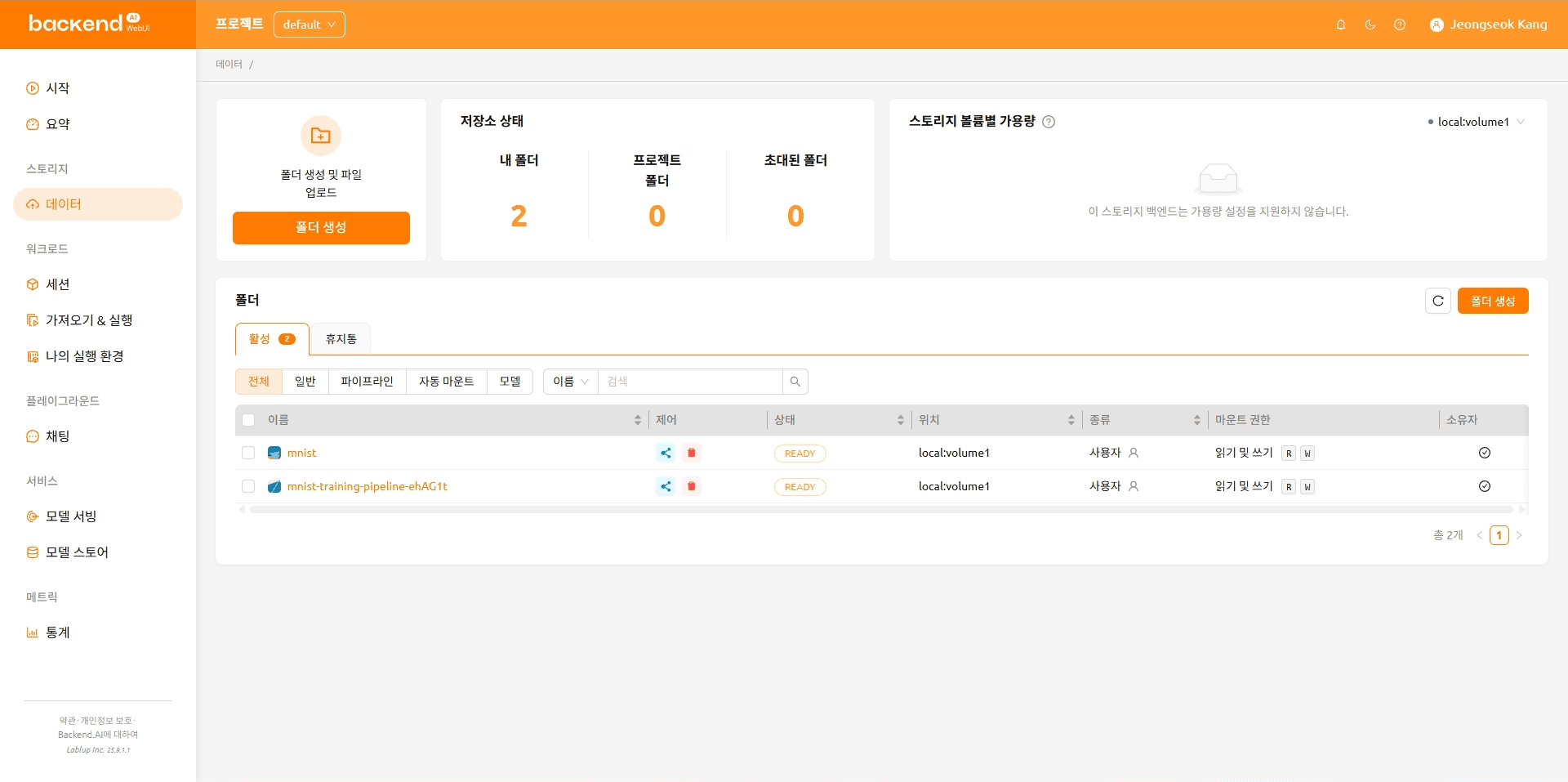
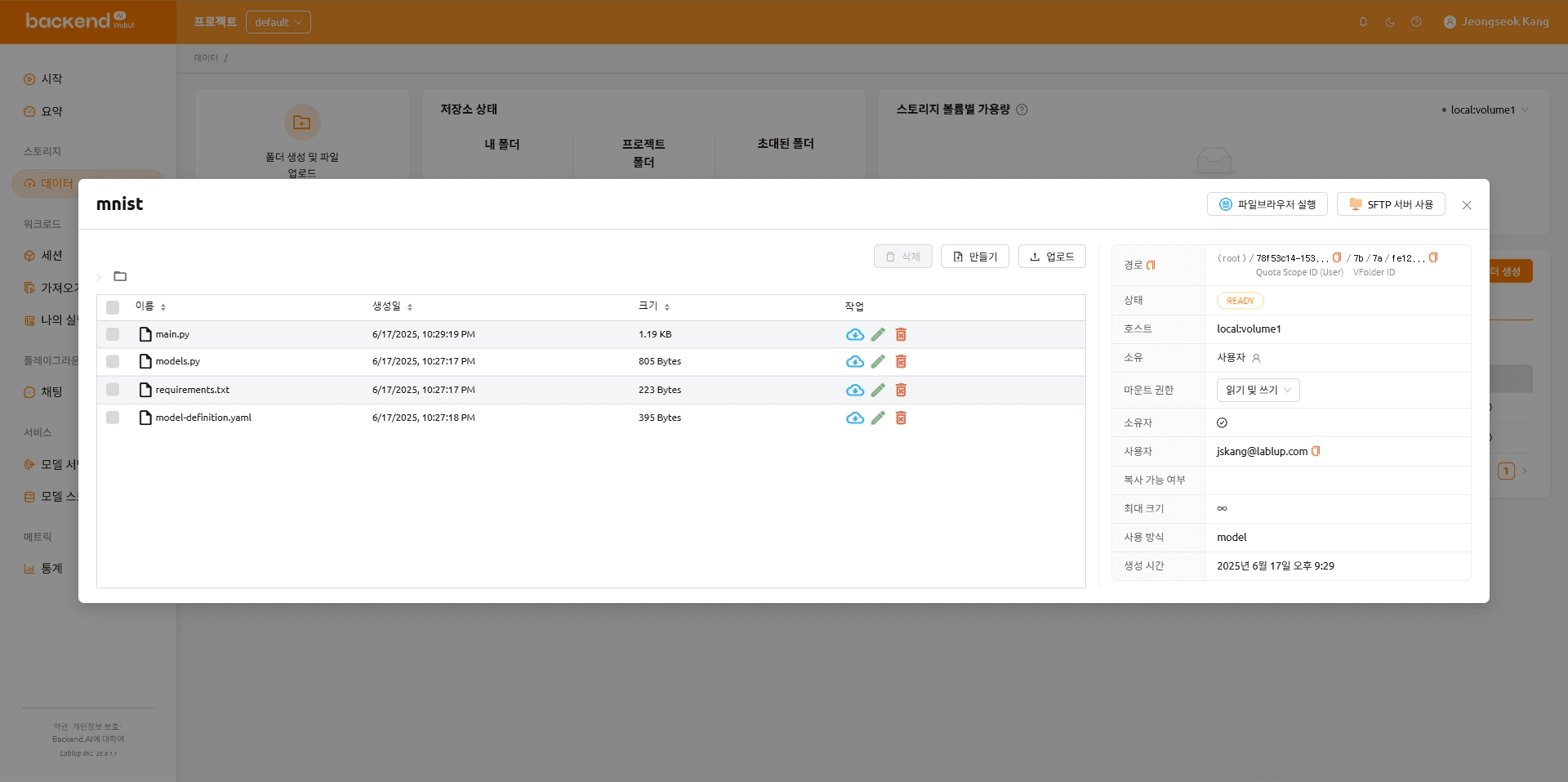
The folder organization after the files are uploaded is shown below.
mnist
|---- main.py
|---- models.py
|---- requirements.txt
\---- model-definition.yaml
import os
from http import HTTPStatus
from pathlib import Path
import gradio
import numpy as np
import torch
from fastapi import FastAPI, Response
from fastapi.responses import JSONResponse
from torchvision import transforms
from models import Net
CHECKPOINT_PATH = Path(os.getenv("CHECKPOINT_PATH")).joinpath(os.getenv("BACKENDAI_PIPELINE_JOB_ID", "mnist_cnn")).with_suffix(".ckpt")
model = Net()
model.load_state_dict(torch.load(CHECKPOINT_PATH))
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,)),
transforms.Grayscale(),
])
app = FastAPI()
@app.get("/health", status_code=HTTPStatus.OK)
async def health_check() -> Response:
return JSONResponse({"healthy": True})
def fn(img: np.ndarray) -> int:
x = transform(img).unsqueeze(0) # Add batch dimension
x = model(x)
x = x.argmax(dim=1, keepdim=True) # Get the index of the max log-probability
return int(x.item()) # Convert tensor to int and return
gradio_app = gradio.Interface(
fn=fn,
inputs=["image"],
outputs=["number"],
)
app = gradio.mount_gradio_app(app, gradio_app, path="/")import torch
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
self.conv2 = nn.Conv2d(32, 64, 3, 1)
self.dropout1 = nn.Dropout(0.25)
self.dropout2 = nn.Dropout(0.5)
self.fc1 = nn.Linear(9216, 128)
self.fc2 = nn.Linear(128, 10)
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = F.max_pool2d(x, 2)
x = self.dropout1(x)
x = torch.flatten(x, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=1)
return outputfastapi[standard]==0.115.12 # https://github.com/fastapi/fastapi
gradio==5.33.0 # https://github.com/gradio-app/gradio
torch==2.7.1 # https://github.com/pytorch/pytorch
torchvision==0.22.1 # https://github.com/pytorch/visionmodels:
- name: mnist-gradio-demo
model_path: /models
service:
pre_start_actions:
- action: run_command
args:
command: ["pip", "install", "-r", "/models/requirements.txt"]
start_command:
- fastapi
- run
- /models/main.py
port: 8000
health_check:
path: /health
max_retries: 5Adding a task
Back in Backend.AI FastTrack 2, add tasks to the pipeline in order.
You can drag and drop the desired task from the list of task templates as shown below. For more information, see the “Creating a task” topic in our previous blog.
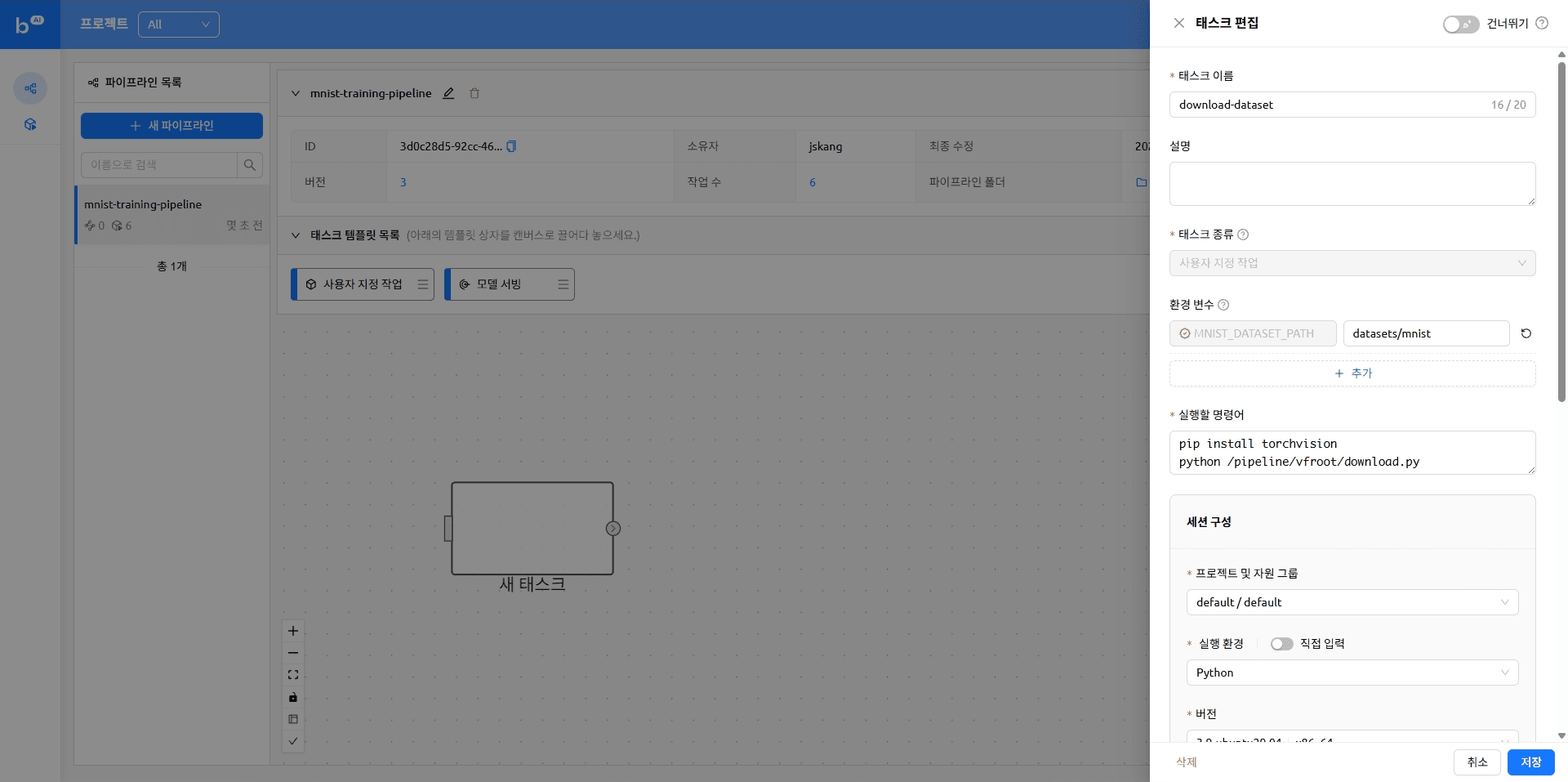 Dragging tasks
Dragging tasks

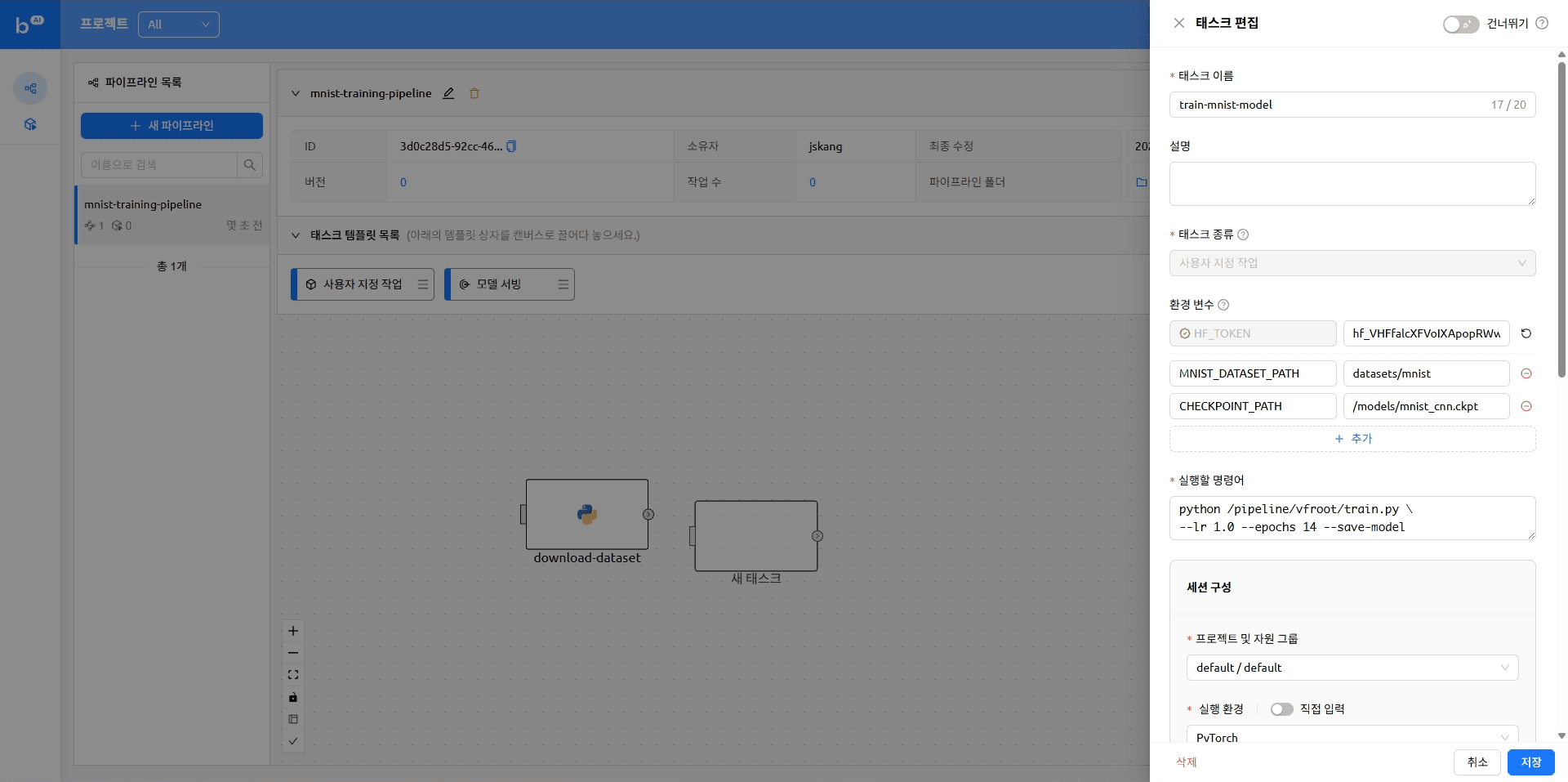
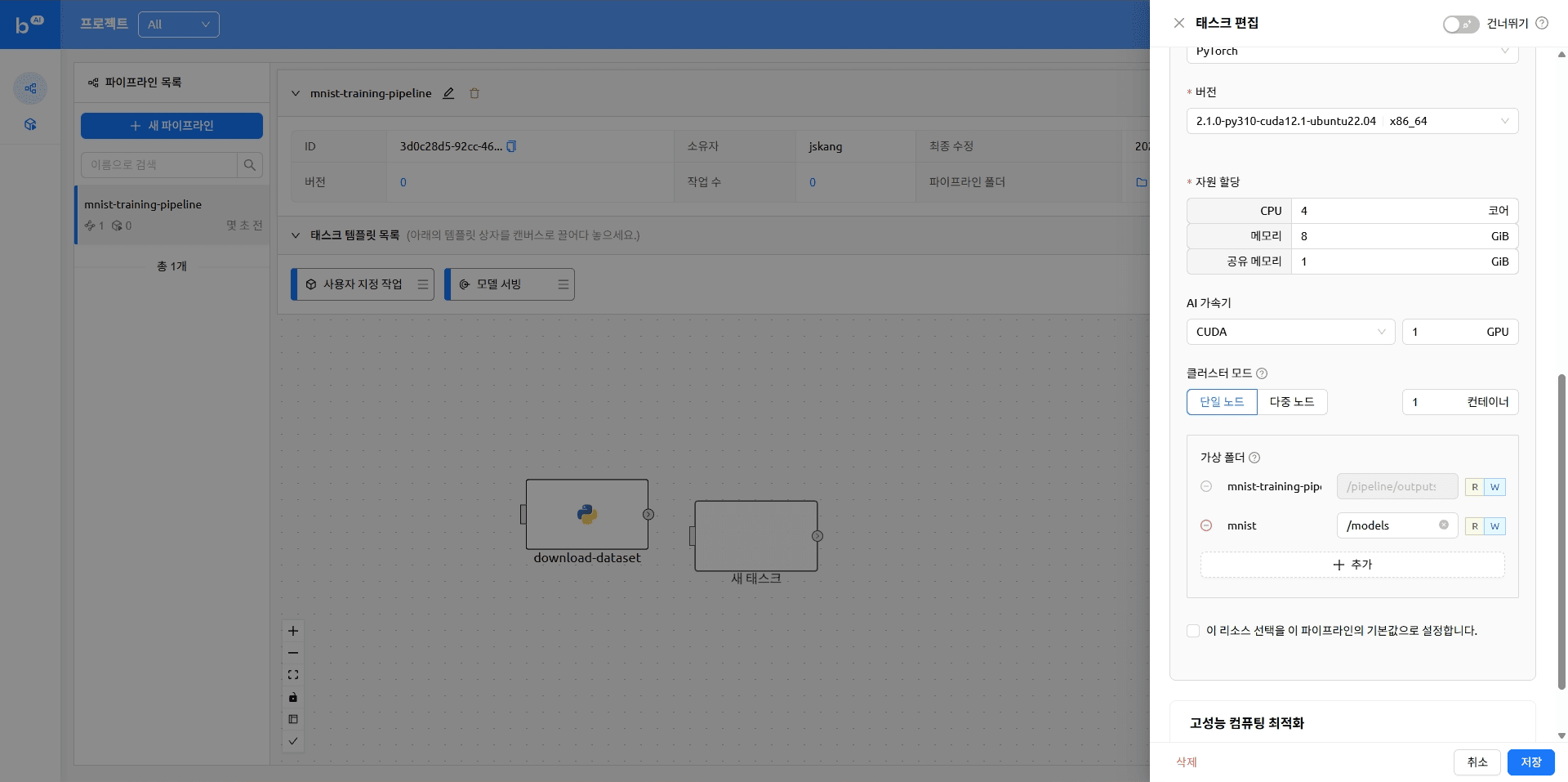
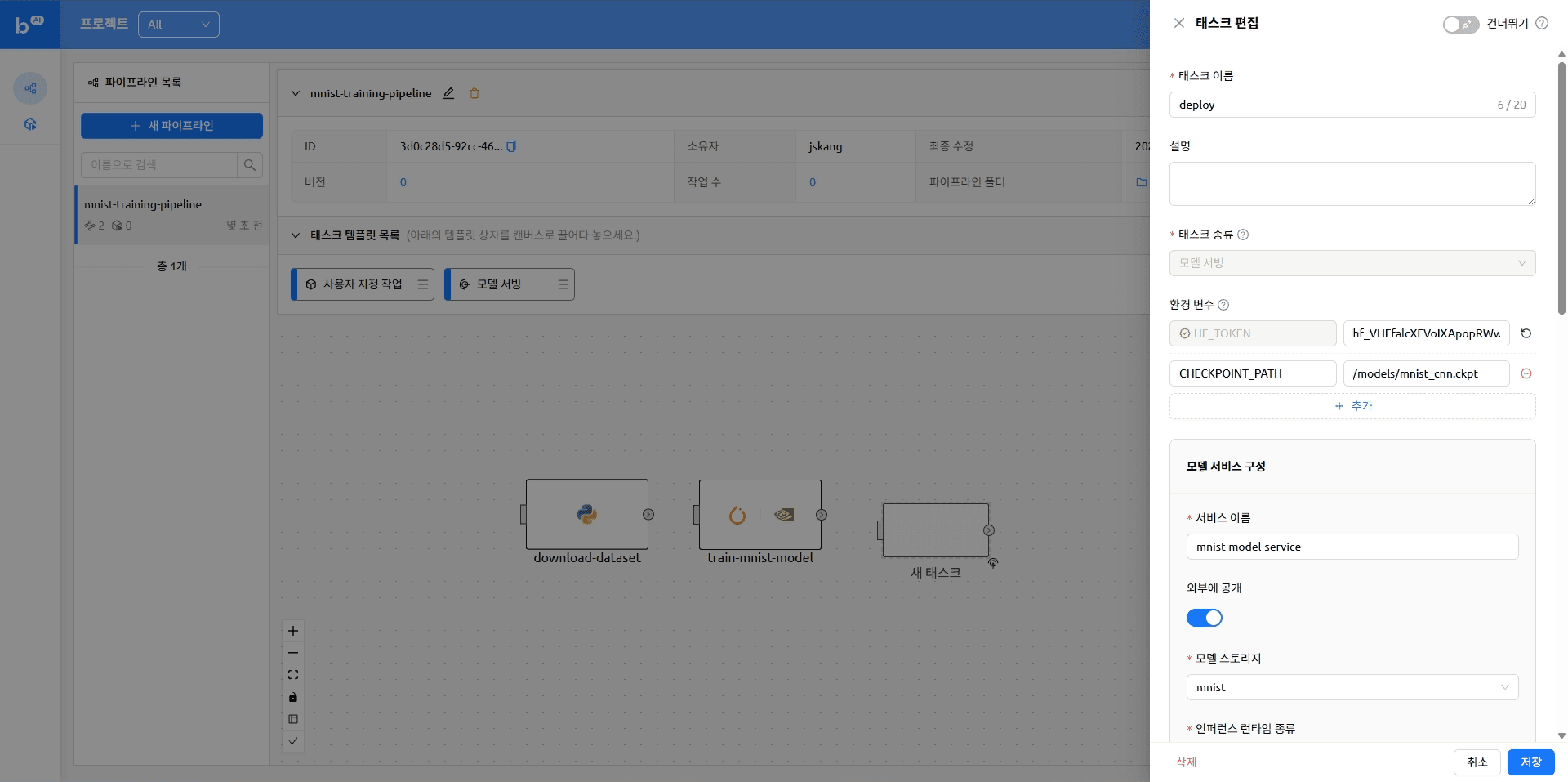
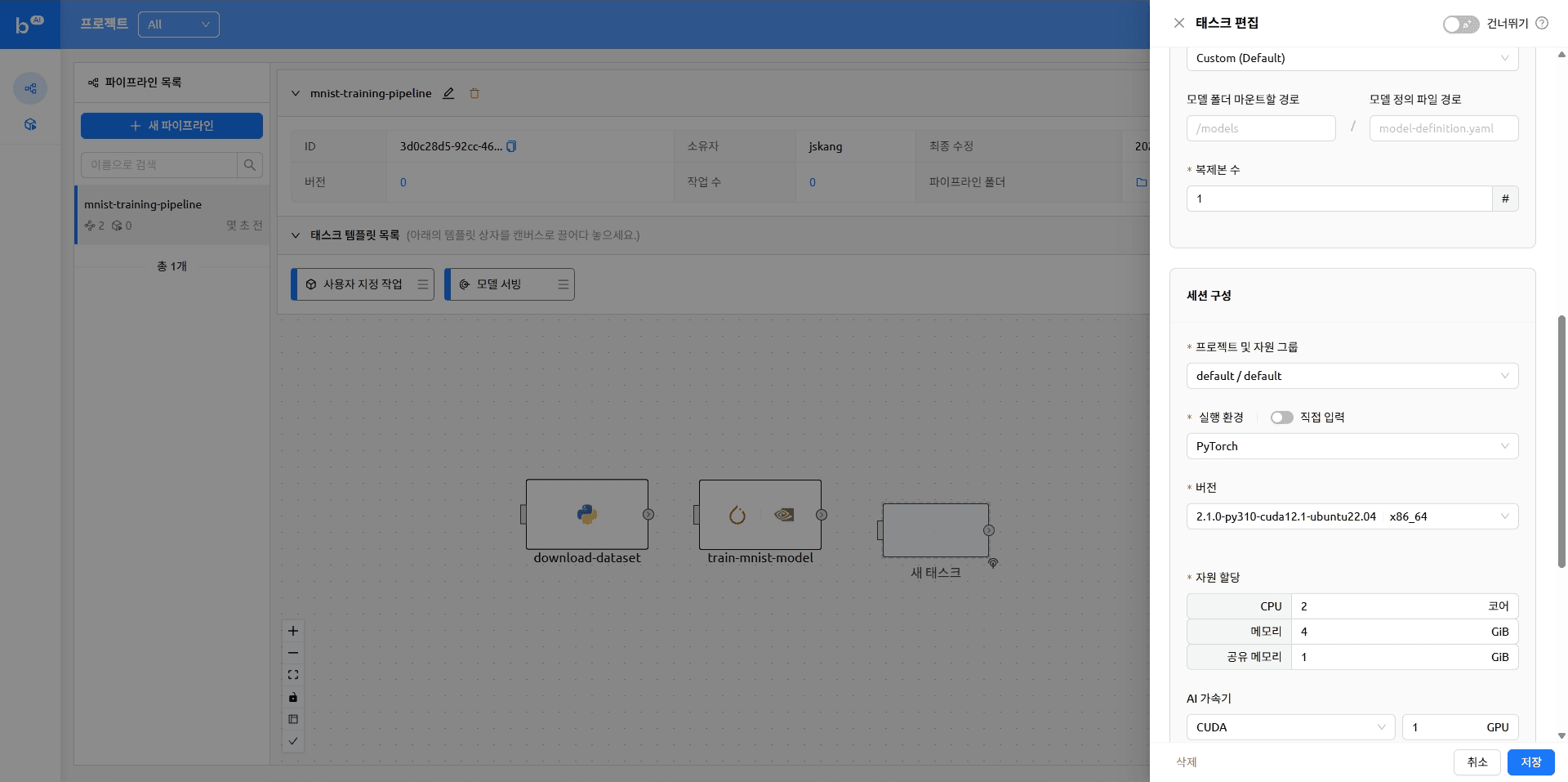
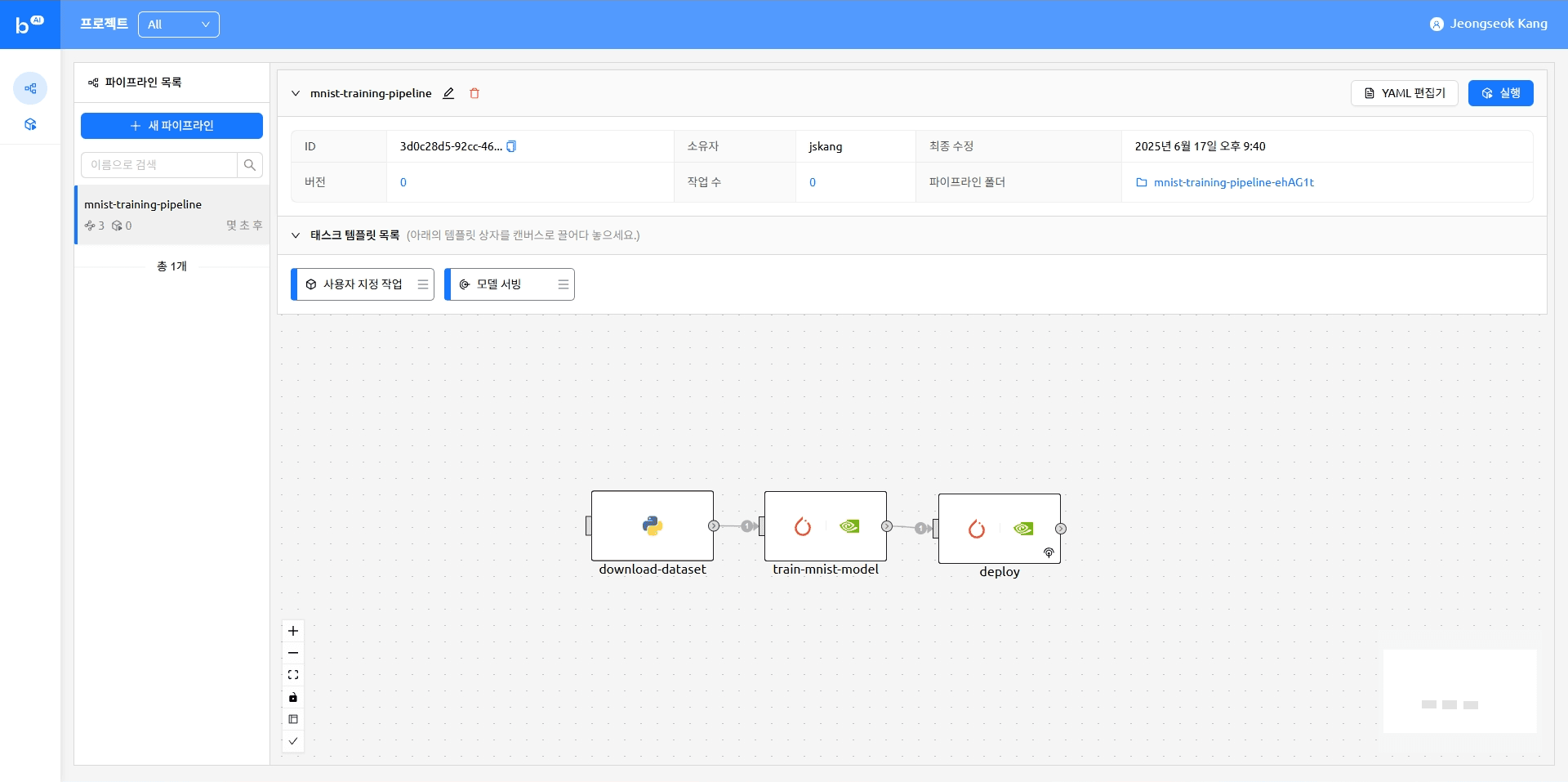
The pipeline with tasks added looks like this
version: 25.9.0
name: mnist-training-pipeline
description: ''
ownership:
domain_name: default
scope: user
environment:
envs:
HF_TOKEN: ****
tasks:
- name: train-mnist-model
description: ''
type: default
cluster_mode: single-node
cluster_size: 1
module_uri: ''
environment:
project: default
scaling-group: default
image: cr.backend.ai/stable/python-pytorch:2.1.0-py310-cuda12.1-ubuntu22.04
envs:
MNIST_DATASET_PATH: datasets/mnist
CHECKPOINT_PATH: /models/mnist_cnn.ckpt
resources:
cpu: 4
mem: 4g
cuda.device: '1'
resource_opts:
shmem: 0g
dependencies:
- download-dataset
mounts:
- mnist:/models
skip: false
command: python /pipeline/vfroot/mnist/train.py --lr 1.0 --epochs 14 --save-model
- name: download-dataset
description: ''
type: default
cluster_mode: single-node
cluster_size: 1
module_uri: ''
environment:
project: default
scaling-group: default
image: cr.backend.ai/stable/python:3.9-ubuntu20.04
envs:
MNIST_DATASET_PATH: datasets/mnist
resources:
cpu: 1
mem: 4g
resource_opts:
shmem: 0g
dependencies: []
mounts: []
skip: false
command: 'pip install torchvision
python /pipeline/vfroot/mnist/download.py'
- name: deploy
description: ''
type: serving
cluster_mode: single-node
cluster_size: 1
module_uri: ''
environment:
project: default
scaling-group: default
image: cr.backend.ai/stable/python-pytorch:2.1.0-py310-cuda12.1-ubuntu22.04
envs: {}
resources:
cpu: 2
mem: 4g
cuda.device: '1'
resource_opts:
shmem: 0g
dependencies:
- train-mnist-model
mounts: []
skip: false
service:
name: mnist
model: mnist
model_mount_destination: /models
runtime_variant: custom
replicas: 1
open_to_public: trueRunning a Pipeline
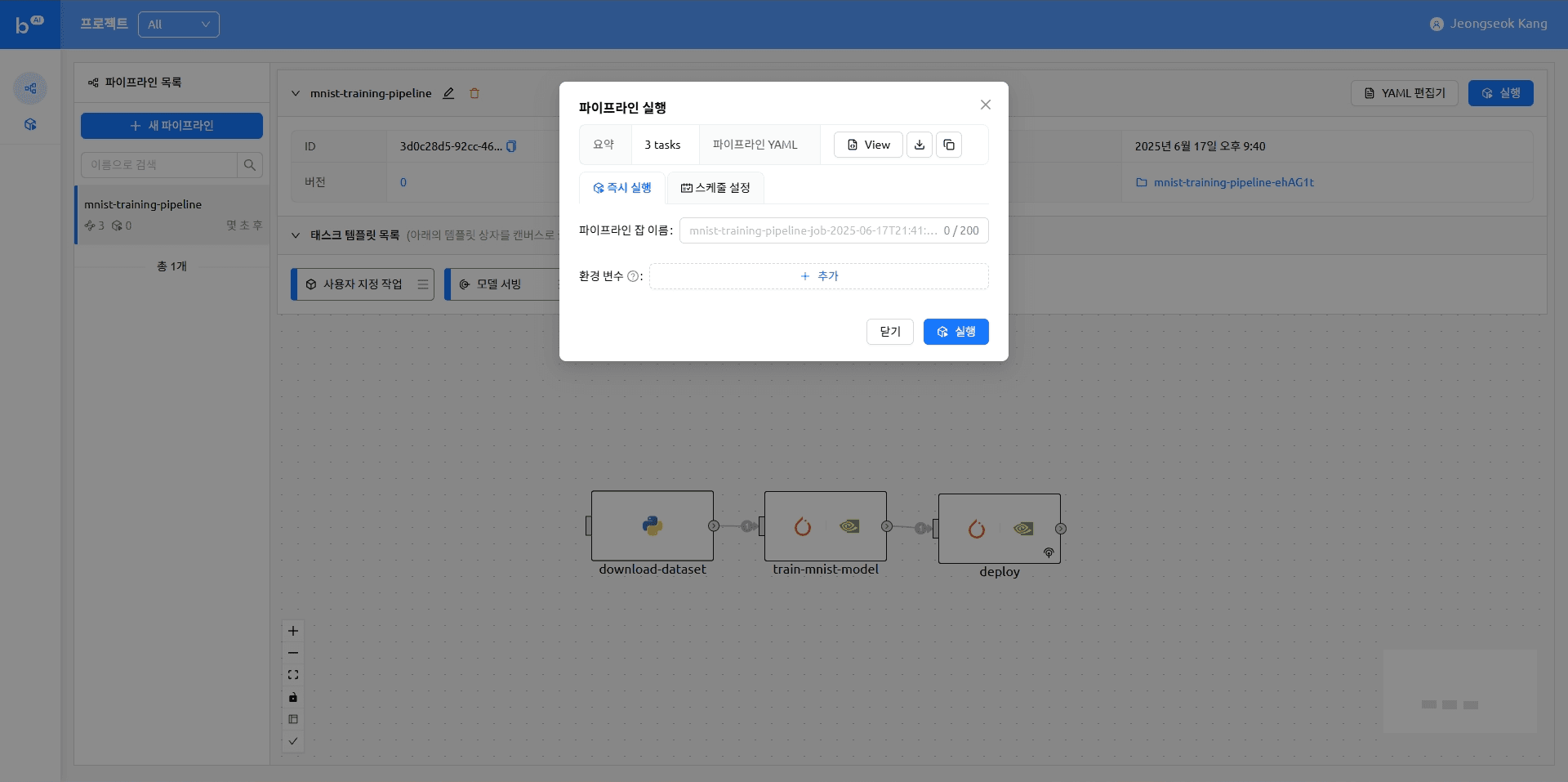
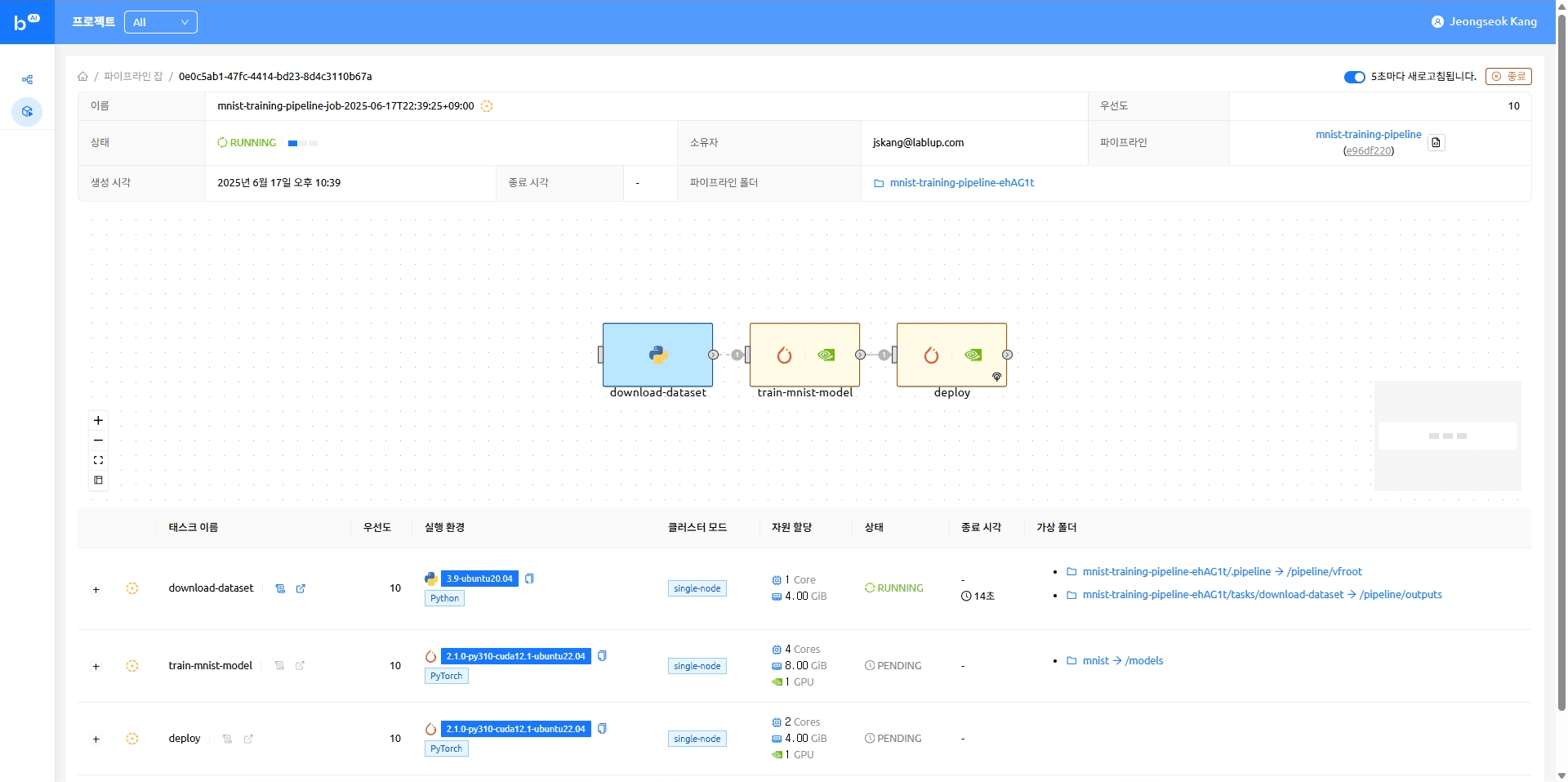
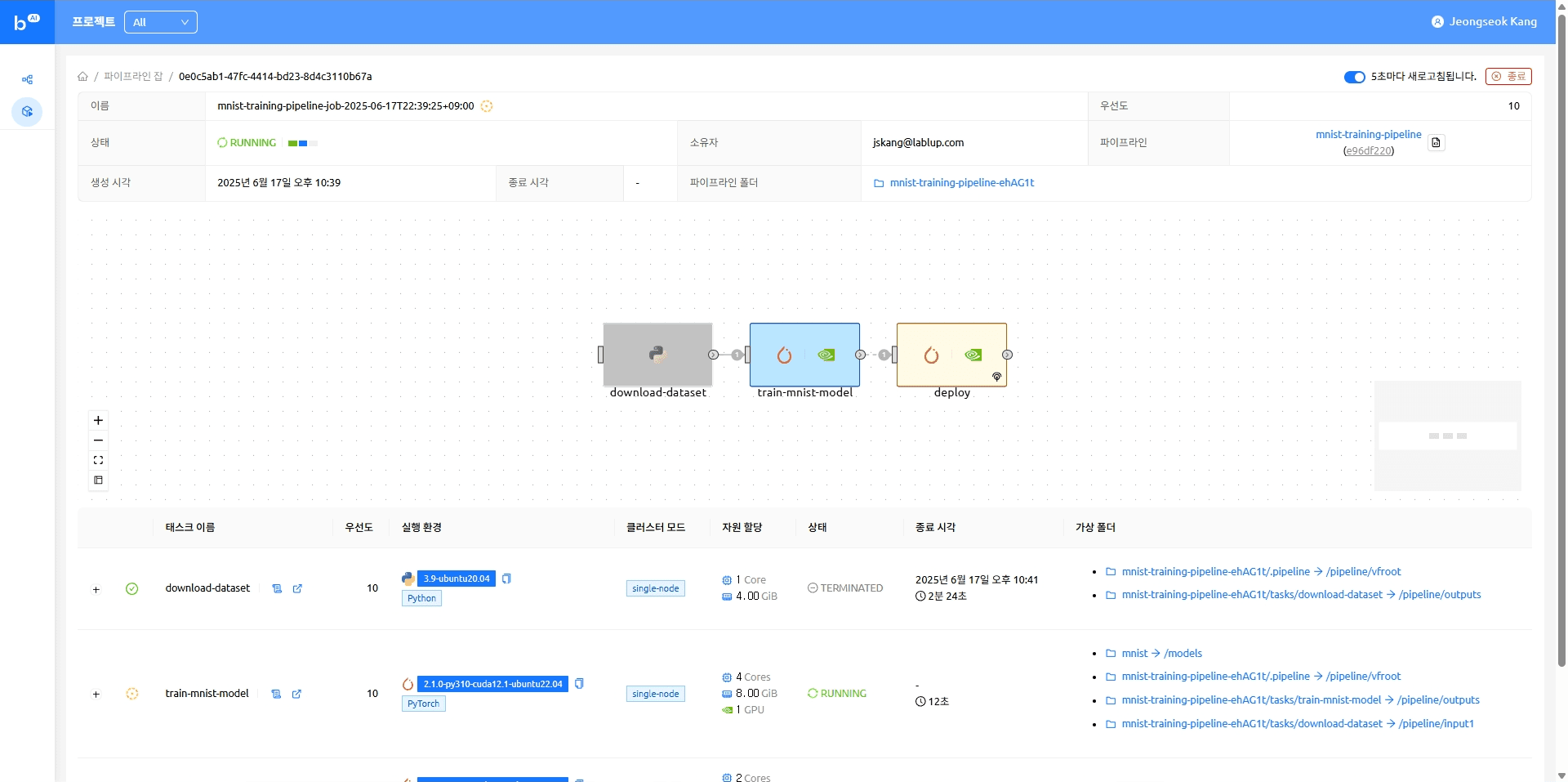
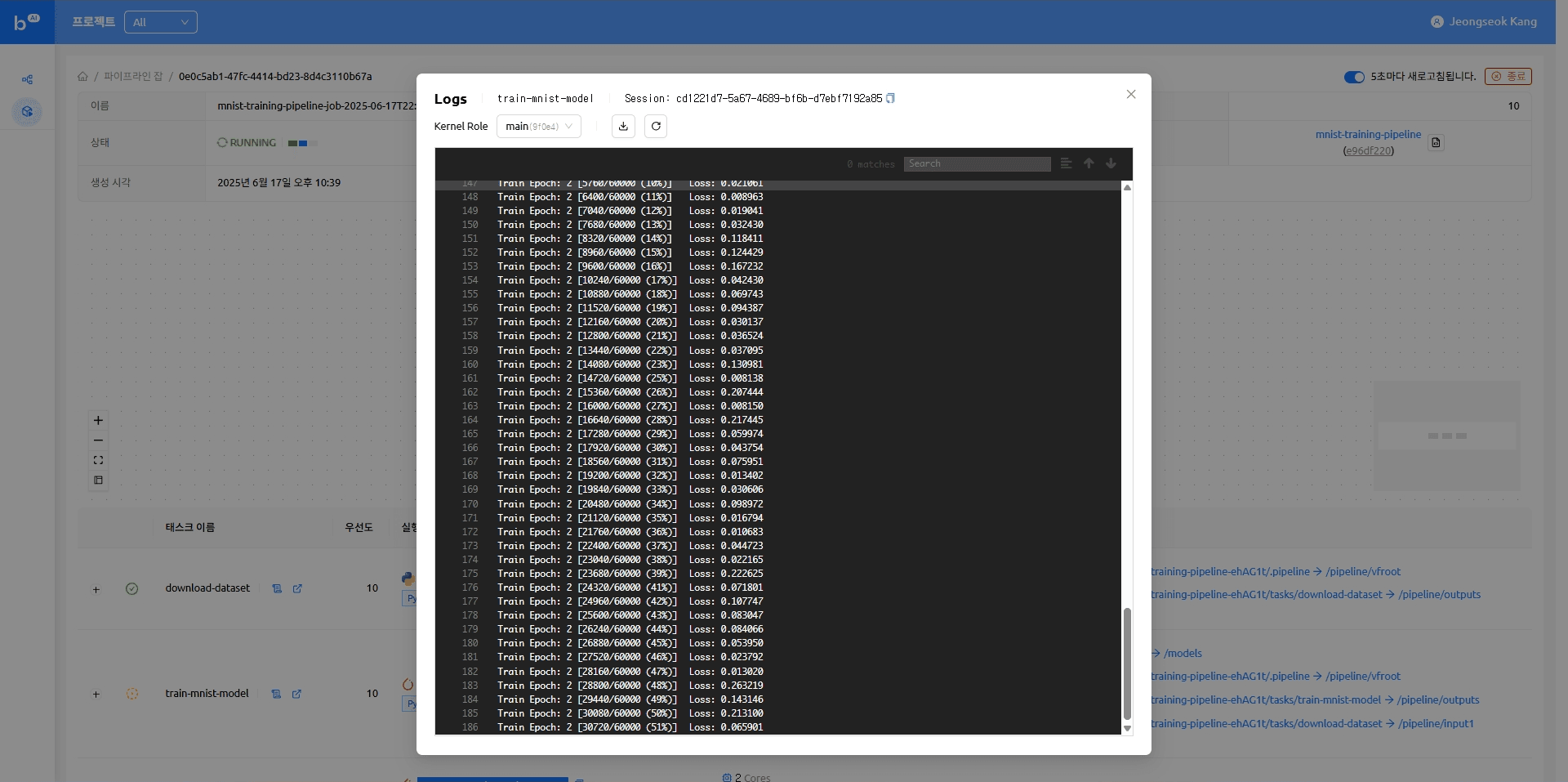
When the model serving task completes, the model appears as an accessible endpoint. Clicking that endpoint takes you to the Gradio demo page where you can try running model inference.
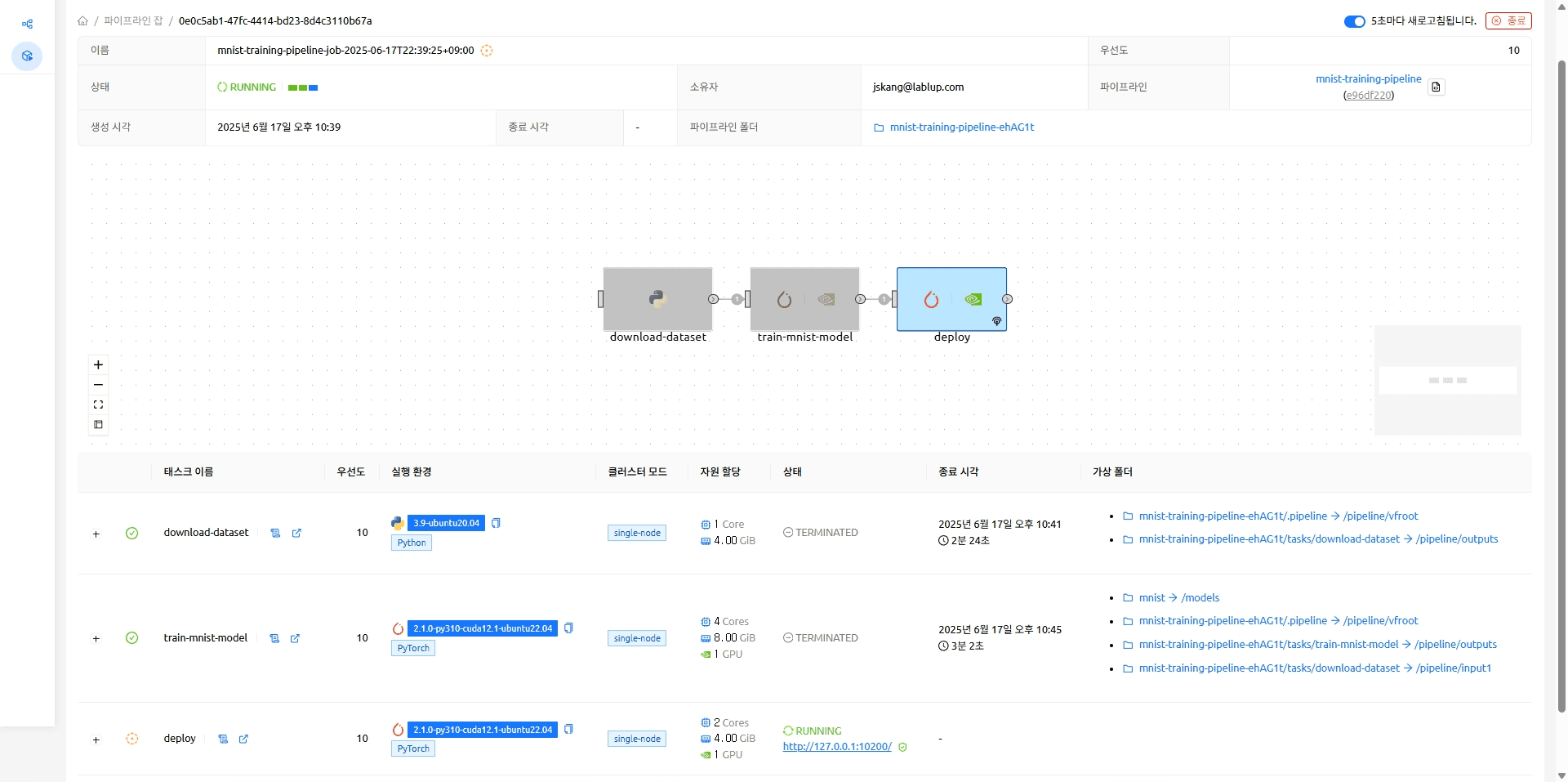 The endpoint on which the MNIST model is running (http://127.0.0.1:10200)
The endpoint on which the MNIST model is running (http://127.0.0.1:10200)
Serving a Model
Upload some random MNIST data and run inference to see that the model trained well.
Closing
In this post, we showed you how to build a simple model training and serving pipeline using Backend.AI FastTrack 2. As always, we're always interested in hearing from machine learning researchers. If there's a topic you'd like to see us cover further, feel free to send us your thoughts at contact@lablup.com.
Thank You!
Appendix
MNIST Dataset
You can use the Python script below to convert a MNIST dataset file of the form MNIST/raw/t10k-images-idx3-ubyte to an image file.
import json
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
root_path = Path(__file__).parent.parent
with open(root_path / "datasets/mnist/train/dataset_info.json", "r") as f:
dataset_info = json.load(f)
image_size = 28
test_num_examples = dataset_info["splits"]["test"]["num_examples"]
with open(root_path / "datasets/mnist/MNIST/raw/t10k-images-idx3-ubyte", "rb") as f:
f.read(16) # Skip the header
buf = f.read(image_size * image_size * test_num_examples)
data = np.frombuffer(buf, dtype=np.uint8).astype(np.float32)
data = data.reshape(test_num_examples, image_size, image_size)
for i in range(10):
plt.imsave(root_path / f"datasets/{i}.png", data[i], cmap="gray")Footnotes
-
Sculley, David, et al. "Hidden technical debt in machine learning systems." Advances in neural information processing systems 28 (2015). ↩
-
Jimmy Lin and Dmitriy Ryaboy. 2013. Scaling big data mining infrastructure: the twitter experience. SIGKDD Explor. Newsl. 14, 2 (December 2012), 6–19. https://doi.org/10.1145/2481244.2481247 ↩
-
D. Kreuzberger, N. Kühl and S. Hirschl, "Machine Learning Operations (MLOps): Overview, Definition, and Architecture," in IEEE Access, vol. 11, pp. 31866-31879, 2023, doi: 10.1109/ACCESS.2023.3262138. ↩





